CRISPR in Nature
Introduction
WHERE DID CRISPR COME FROM?
CRISPR systems evolved to defend bacteria against bacterial viruses called phages. Learn how phages infect bacteria and how those bacteria use CRISPR and other defenses to fight back.
NOTE: On this page, we use the term bacteria instead of the correct, more general term prokaryotes, since people are more familiar with bacteria. Please note, however, that the general information described here applies to both bacteria and the abundant but lesser-known archaea.
The phage life cycle
How phages infect bacteria and spread
VOCABULARY
Phage, bacteriophage, phage tail, phage tail fibers, phage head, lyse, lytic cycle, lysogenic
WHAT IS A PHAGE?
The word phage is short for bacteriophage, the type of virus that infects bacteria.
Deep in the ocean, in our own gut, and everywhere in between, phages are infecting all sorts of bacteria. Even though they’re invisible to the naked eye, phages are actually the most abundant life-like form on Earth! We say “life-like” because viruses do not meet the criteria to be considered living organisms.
Just as human cells use DNA as instructions for their function, phages carry around DNA (or RNA) genomes as blueprints for their assembly and spread. However, phages cannot reproduce on their own. They must take resources from bacteria during the process of infection and use these resources to produce more phages. This is why they are only “life-like.”
THE PHAGE INFECTION PROCESS

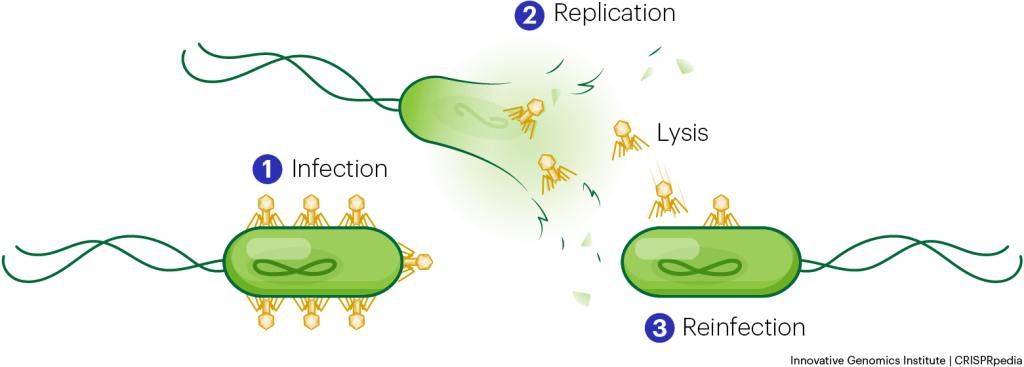
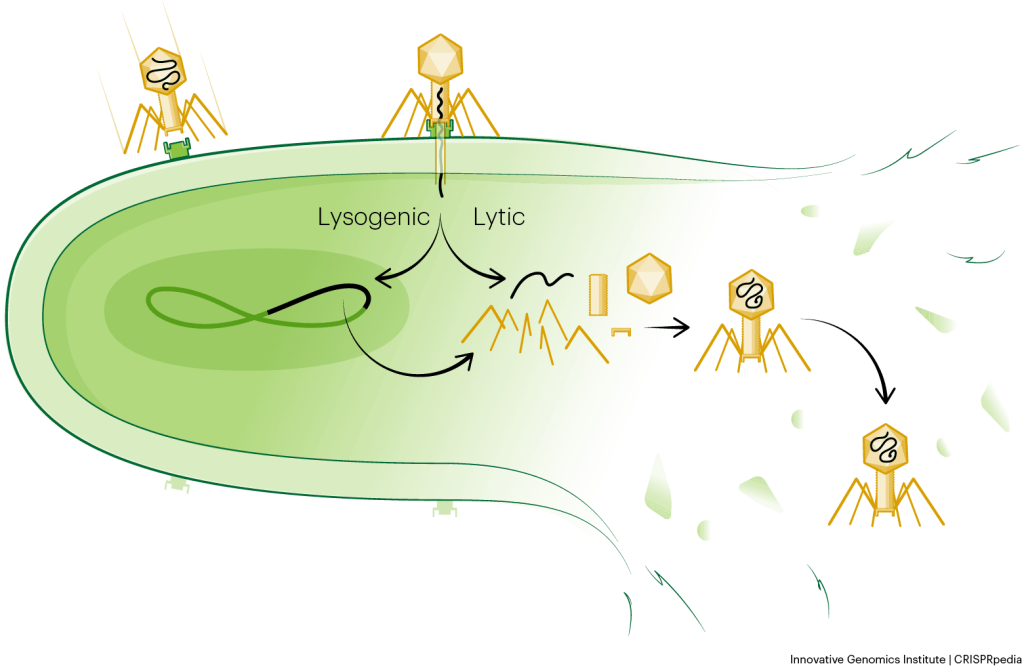
With the aid of a microscope, scientists can watch the phage infection process in action. Phages look like tiny moon-landers (but they can have other interesting shapes too!). Their curious structures typically include pieces that resemble a head, neck, and legs. When a phage lands on a bacterium, it “locks onto” the bacterial surface using its legs or tail fibers. Then, the phage’s DNA travels from storage space in its head, down a long tube called the tail, and into the bacterial cell.
After a phage injects its DNA into a bacterium, the bacterium’s own protein machinery replicates the phage genome. The bacterium then follows the instructions in the phage DNA and generates more heads, tails, tail fibers, and other phage pieces. These “body parts” assemble to form new phages.
Once the phages reach a critical mass, they explode out of or lyse the bacterium and escape into the environment. This is similar to a balloon popping.
Each time a new bacterial cell is infected, a new swarm of phages is produced. The infection can thereby spread exponentially, quickly taking over communities of bacteria. This is known as the lytic cycle. In contrast, so-called lysogenic phages hide their DNA inside an infected bacterium’s genome. They lie in wait and only produce more phages at a later time when conditions are right.
Bacterial defenses
The many ways that bacteria defend themselves against invasive DNA
VOCABULARY
Bacterial receptor, cytoplasm, capsid, restriction modification, restriction enzymes, methyl groups, innate defenses, adaptive immunity
HOW DO BACTERIA DEFEND AGAINST VIRUSES?
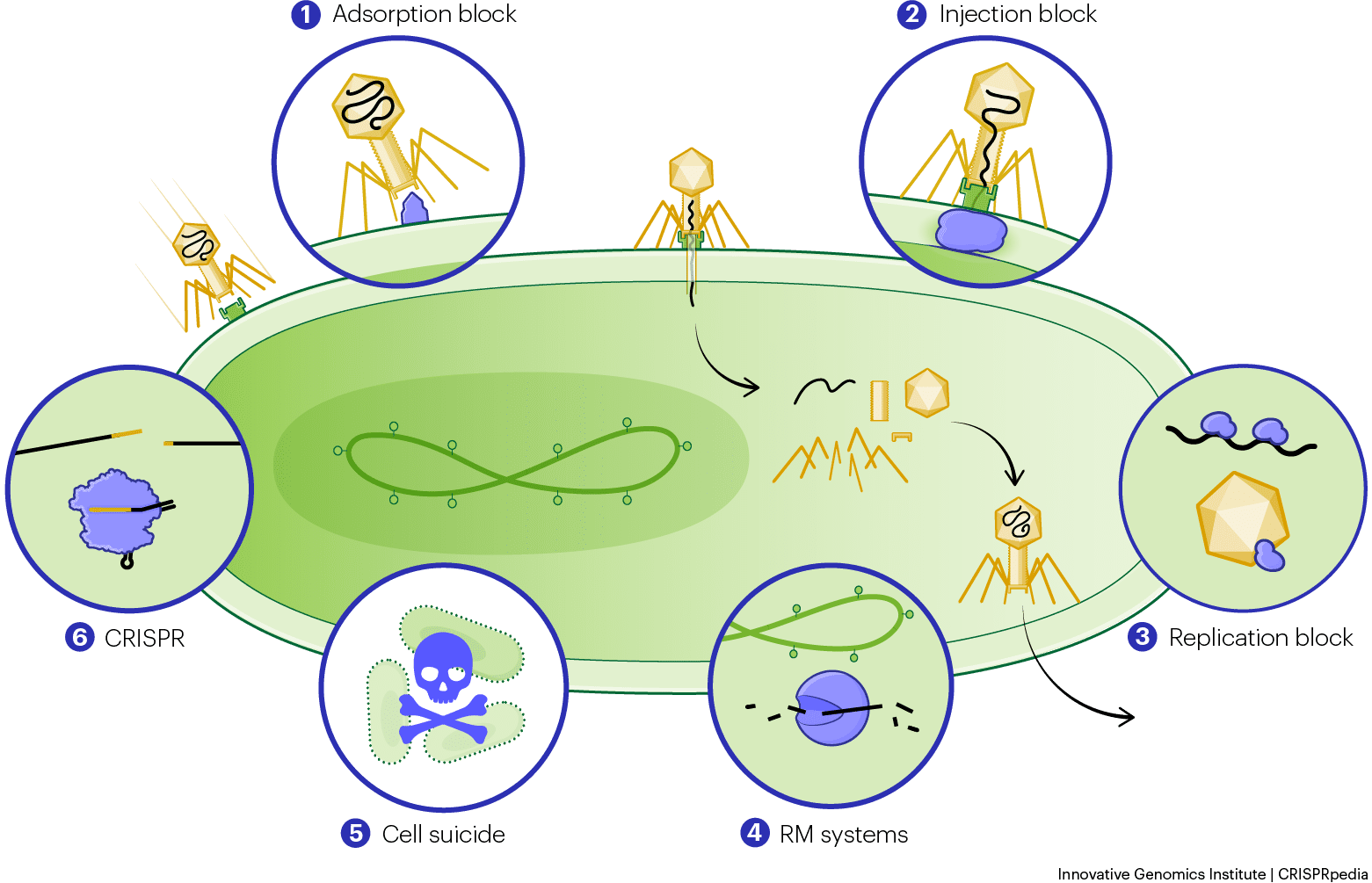
In nature, phages outnumber bacteria ten to one. If phages are so good at killing bacteria, how are there any bacteria left on Earth? Bacteria have evolved a whole range of tricks to fend off phages. Each of their defense systems works a bit differently, and we’re only showing some of the general types here. There are many, and we’re still learning how they all work!
PREVENTING ADSORPTION
Perhaps the most obvious defense is to prevent phages from landing on bacterial cells in the first place. The bacterial cell membrane is covered with different receptors, proteins, or groups of proteins that recognize molecules from the outside environment and transmit signals into the bacterium. Phages have taken advantage of bacterial receptors for their own purposes. They attach to their bacterial hosts by sticking to receptors in a process called adsorption. Some bacteria keep phages from binding to their surfaces by altering the structure of their receptors or introducing physical barriers to prevent attachment.
BLOCKING CYTOPLASMIC ENTRY
Alternatively, some bacteria block phage DNA from entering the cytoplasm (the cell interior). In order to inject their DNA into a bacterial cell, phages must penetrate the cell membrane. By adding new proteins into this membrane, bacteria can clog the phage entryway and block DNA injection.
INTERFERING WITH PHAGE REPLICATION
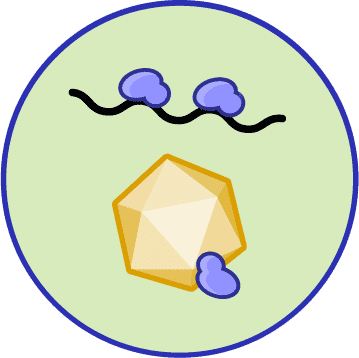
When phage DNA makes it into the cytoplasm, some bacteria can prevent the genome from being replicated, while others prevent replicated phage genomes from being loaded into capsids and bursting out of the cell.
RESTRICTION MODIFICATION SYSTEMS
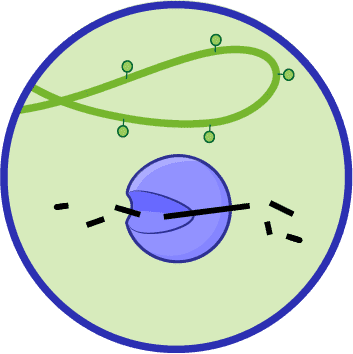
Many bacteria deploy restriction modification (RM) systems to destroy phage DNA that is injected into the cell. These defense systems are composed of scissor-like proteins called restriction enzymes. These enzymes cut phage DNA apart, thereby destroying the instructions for making more phages.
To prevent their own DNA from being damaged by restriction enzymes, bacteria add protective chemicals called methyl groups to their genomes. Restriction enzymes have evolved to ignore methylated DNA and don’t cut it up. Thus, methylation keeps the bacterial genome safe.
CELL SUICIDE

If a phage manages to bypass all these safeguards, the bacterium’s last line of defense is cell suicide. This “altruistic” act kills the individual bacterium, but prevents the production of more phage copies that could go on to infect neighboring cells. One common version of this process is known as abortive infection.
BEYOND INNATE DEFENSES
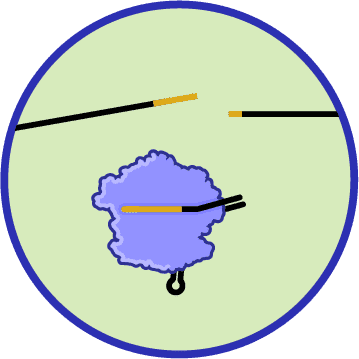
All the defense systems described above are considered innate defenses. This means that they generally evolve slowly, act quickly during infection, and defend against phages in general rather than against any one specific phage. Almost every bacterium has some form of innate defense.
About half of bacteria also have an adaptive immune mechanism called a CRISPR-Cas system, which defends against specific types of phages. This system can adapt, rapidly generating immunity against new phage challengers. There are other types of adaptive immune systems too. We’re focusing on CRISPR-Cas immunity, which we’ll describe in more detail in the next section.
The CRISPR immune system
Bacteria remember and fend off specific phage threats through CRISPR-Cas immunity
VOCABULARY
CRISPR-Cas system, repeats, spacers, Cas proteins, acquisition/adaptation, RNA, RNA biogensis, precursor CRISPR RNA/pre-crRNA, CRISPR RNA/crRNA, interference, complex, search complex/effector complex/ surveillance complex, prespacer, protospacer, complementary
WHAT DO CRISPR SYSTEMS DO IN NATURE?

Bacteria use a variety of defenses to block attacks from phages, including a unique form of adaptive immunity known as a CRISPR-Cas system (or CRISPR system, for short). These systems are found in about half of all bacterial species and nearly all archaea, though they may be less common in microbes that we can’t culture in isolation. In a nutshell, CRISPR systems work by capturing small pieces of invading phage DNA, stashing them in the host cell’s genome, and using these molecular memories to find and destroy matching phages.
So where does CRISPR get its unusual name? The acronym CRISPR (clustered regularly interspaced short palindromic repeats) comes from a section of DNA within the bacterial genome. This DNA is called a CRISPR array or repeat-spacer array, and it has two components — repeats and spacers.
- Repeats — These are segments of DNA that all have the same sequence. They’re generally around 30 base pairs long. The repeats are called “palindromic” in the CRISPR acronym because they usually contain a stretch of bases followed by the complementary bases of the same sequence, in reverse order. It can be hard to envision, so looking at illustrations and visualizations can help. The terminology comes from palindromes in language: words or phrases that can be read the same way forward and backward, like “race car.”
This internal complementarity is important because it means that most CRISPR repeats fold into a structure called a stem-loop or hairpin once they’re transcribed into RNA.
NOTE: The Cas9 system does not have palindromic repeats. Instead of a single crRNA that base-pairs internally, Cas9’s crRNA base-pairs with a second piece of RNA, called the tracrRNA (shown in the first illustration in the next chapter). - Spacers — These sit between the repeats. They are small segments of phage DNA, each one different from the next. These are the “molecular memories” described above. Spacers are remnants from previous phage infections and they get passed down through generations of bacteria. This means that an individual cell doesn’t have to have been infected itself in order to be immune from a phage.
In most cases, you’ll find a set of CRISPR-associated, or cas, genes right next to the repeat-spacer array. These genes encode Cas proteins, which carry out all the activities needed for immunity. Besides fending off phages, CRISPR systems can also block other invasive nucleic acids, like plasmids.
There are a variety of different CRISPR systems, but they all defend bacteria by following three basic steps: spacer acquisition, CRISPR RNA biogenesis, and interference. Let’s go through each step in a little more detail.
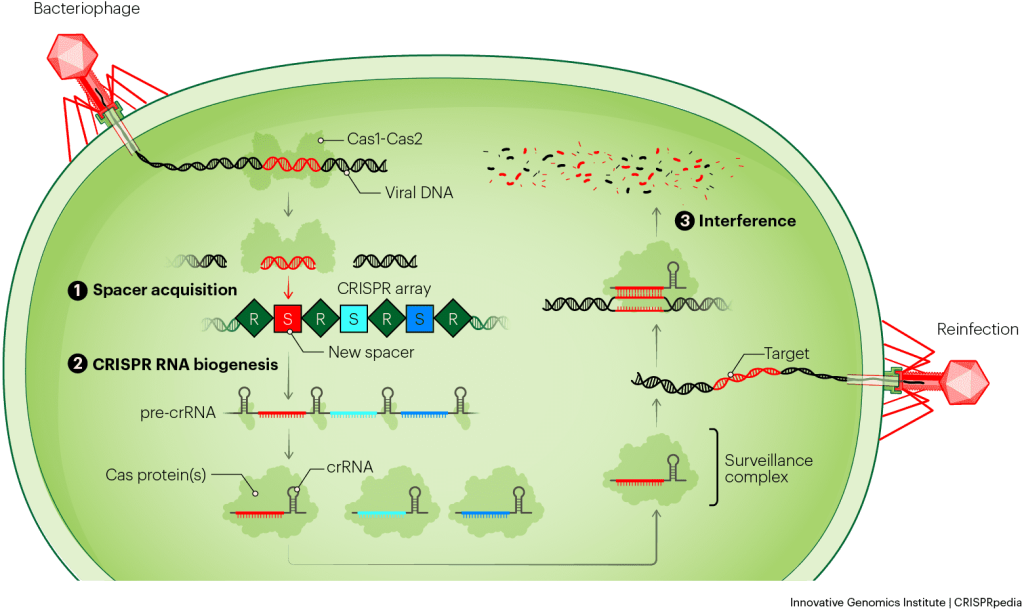
SPACER ACQUISITION
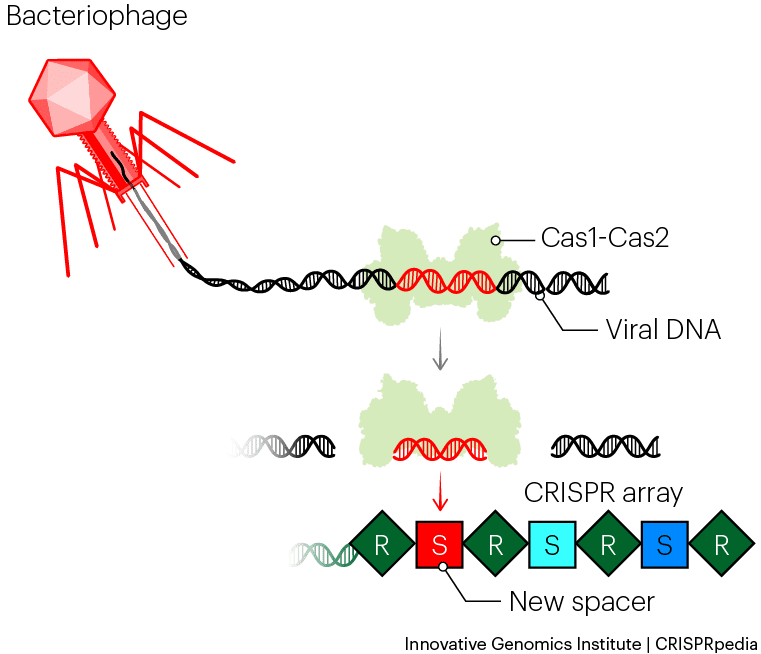
CRISPR systems work by remembering phage infections, so the first step of CRISPR immunity is making a memory. When a phage infects a bacterium equipped with a CRISPR-Cas system, a new spacer sequence can be added to the CRISPR array. This proceeds through a process called acquisition or adaptation.
In this process, two Cas proteins, Cas1 and Cas2, work together to capture pieces of injected phage DNA. The Cas1–Cas2 complex acts as a ruler and works with accessory proteins to measure and cut out a precisely-sized piece of DNA. The complex holds onto the phage DNA and inserts it into one end of the CRISPR array as a new spacer. The system incorporates a new repeat in the process, so all spacers are always flanked by repeats on each side. The bacterium thereby stores a memory of the phage.
This step is akin to a vaccination — you can imagine the repeat-spacer array is like a vaccination card that shows all the phages against which the bacterium is immune.
CRISPR RNA BIOGENESIS
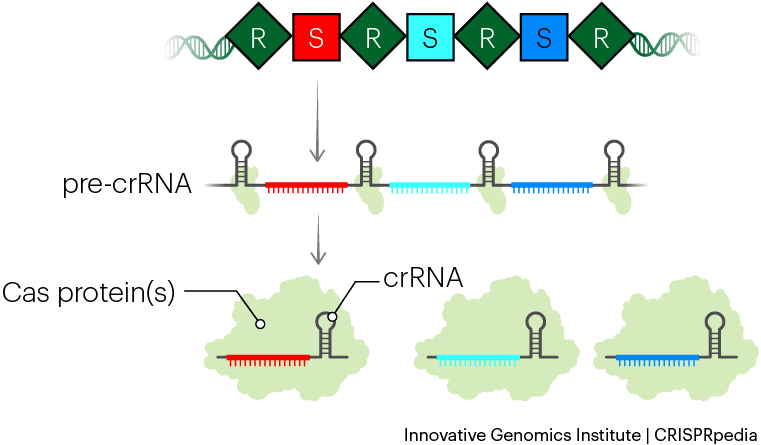
CRISPR systems store these memories of phage infection in their DNA, but don’t use the DNA to recognize subsequent phage infections directly. Instead, they convert the DNA memories into a similar molecule called RNA. RNA is chemically different from DNA and often exists as a single strand. Bacteria make many RNA copies of the phage memories and use them to find newly invading phages. It is useful to make these RNA “working copies” because they can be broken down and recycled without destroying the original, permanent memory.
The biological term for this process is CRISPR RNA biogenesis. It involves transcribing the whole CRISPR array of repeats and spacers into a long piece of RNA called a precursor CRISPR RNA (pre-crRNA). Typically, Cas proteins then cut or ‘cleave’ the long RNA into short, individual segments. These contain one spacer and parts of the repeats. The final, fully trimmed RNA copies of the phage memories are called CRISPR RNAs (crRNAs). Next, Cas proteins will use each crRNA to hunt for and positively identify matching phage DNA.
INTERFERENCE
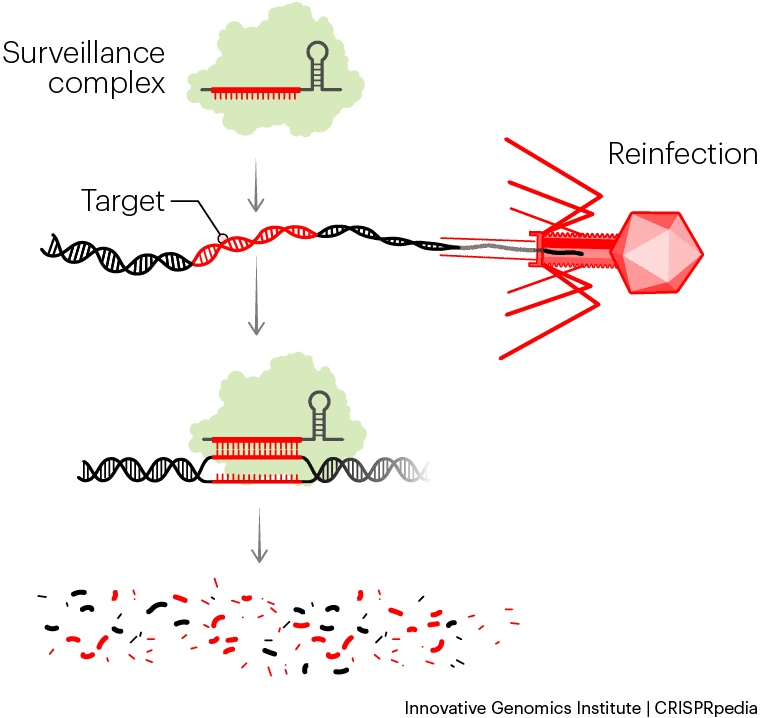
The final step in CRISPR immunity leads to the destruction of invading phage DNA. This process is called interference, because it involves the CRISPR system “interfering with” or stopping the phage’s life cycle. Interference starts with a single Cas protein or a group of Cas proteins grabbing onto or binding one crRNA. Together, the Cas protein(s) plus crRNA form what is known as the search complex, effector complex, or what we’ll call it here — the surveillance complex. (When multiple molecules of protein, RNA, or other components stick to each other and work together, they’re called a complex.)
When a phage injects its DNA into a bacterial cell, the surveillance complex scouts the phage’s genome for a sequence that matches the spacer in its crRNA. This target sequence is called the protospacer. Because the crRNA is a copy of the original phage DNA, it is complementary to, and can form base pairs with, one strand of the DNA injected by another phage of the same species. Base pairs between RNA and DNA look very similar to the ones between each strand of DNA in a normal double helix.
The surveillance complex unzips the phage DNA and checks to see if the crRNA can base-pair with one strand. If the whole spacer can base-pair, it means the surveillance complex has found the target it’s been searching for, so it cleaves the phage DNA to destroy it. If the surveillance complex searches through all the DNA in the cell but doesn’t find a match, it doesn’t make any cuts.
CRISPR surveillance complexes destroy DNA by cutting it apart. They make different kinds of cuts depending on the CRISPR-Cas system. Some systems make a single break in each strand. Some cut both the target and any nearby RNA or DNA. Still others chew the target up bit by bit, like a lawnmower. After the phage DNA is cut at least once, other proteins inside the cell help break down the rest.
Since the phage’s replication instructions are destroyed, the infection is over. Thanks to its CRISPR defense system, the bacterium survives. In summary, the CRISPR system protected the cell by remembering an old phage infection and using that memory to recognize and stop the same type of phage when it attacked again.
🎥 | You can review the entire CRISPR immunity process in the video below. This animation, created for IGI by Janet Iwasa, depicts a Cas9-containing CRISPR system as an example, so it’s a little more specific than the overview we’ve provided here, but you’ll see all three key steps: spacer acquisition, crRNA biogenesis, and interference.
Reimagining the “CRISPR” acronym
Now that we’ve run through the process of CRISPR-Cas immunity, how might you remember all the steps? We suggest an alternative acronym for CRISPR that might be more useful than “clustered regularly interspaced short palindromic repeats.”
Capture spacer
RNAs transcribed
Interference complex formed
Search for match
Phage genome destroyed
Repeat!
Finally, a note on the potentially confusing variations of ‘spacer’ terminology:
- Prespacer – The acquisition machinery selects a segment of phage DNA called a prespacer and integrates it into the bacterial CRISPR array.
- Spacer – Once the phage DNA is incorporated into the CRISPR array, it is called a spacer. After a crRNA is transcribed from the array, the phage-derived sequence of the RNA is also called a spacer.
- Protospacer – The stretch of DNA in the phage genome that is complementary to the crRNA and cleaved by the surveillance complex is called the protospacer. The PAM is a protospacer-adjacent motif, meaning a short sequence right next to the surveillance complex’s target.
Avoiding self-targeting via the PAM
The protospacer adjacent motif (PAM) prevents CRISPR enzymes from cutting the repeat-spacer array
VOCABULARY
Protospacer adjacent motifs/PAMs
WHAT IS A PAM AND WHY IS IT IMPORTANT?
During CRISPR defense, bacteria use Cas proteins to destroy invading phage DNA. They identify that phage DNA by comparing it to RNA copies of the same sequence stored in their genome’s CRISPR array. Thus, you might be wondering, why don’t Cas proteins end up cutting the spacers in the array?
Recognizing the difference between “self” DNA (the bacterial genome) and “non-self” DNA (foreign DNA like a phage genome) is essential because bacteria typically die when their genomes are damaged. CRISPR systems are able to avoid harmful destruction of the bacterium’s own genome by relying on short DNA sequences called protospacer adjacent motifs, or PAMs. These are found next to CRISPR targets in phage DNA but never in bacterial CRISPR arrays. As a result, Cas proteins cut PAM-containing, phage DNA, but not PAM-less bacterial DNA.
We’ll explain this in more detail using the CRISPR-Cas9 system as an example.
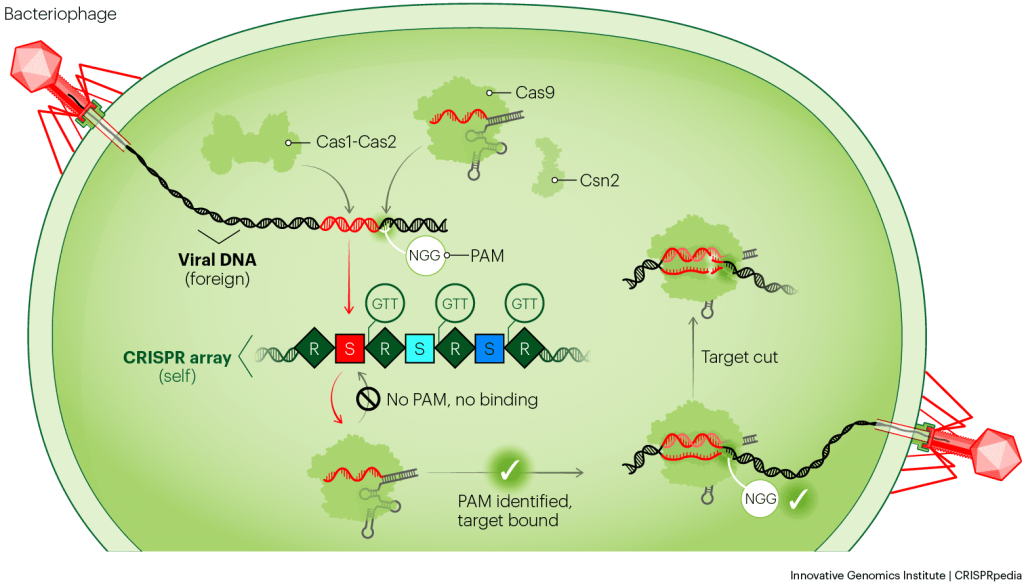
THE CRISPR-CAS9 SYSTEM AND ITS PAM
Cas9 works by searching for any DNA that is complementary to its CRISPR RNA (crRNA) guide. Once Cas9 finds such DNA, it cuts the DNA apart. Since the bacterial CRISPR array encodes the crRNAs, they are always going to be complementary. The PAM requirement is what prevents Cas9 from cutting the array. But how?
Before testing whether its crRNA can base-pair with a stretch of DNA, Cas9 first checks for its specific PAM sequence: GG, two guanine bases. If there’s no PAM, Cas9 floats away. Even if the crRNA contains a perfect, complementary match to a DNA sequence, Cas9 will not find it without first seeing a PAM. When Cas9 successfully finds a PAM, it then checks to see if the adjacent DNA matches its crRNA guide. If it does, then Cas9 knows it’s okay to cut. The repeat-spacer array does not contain PAM sequences, so Cas9 does not cut it.
The SpyCas9 PAM is NGG
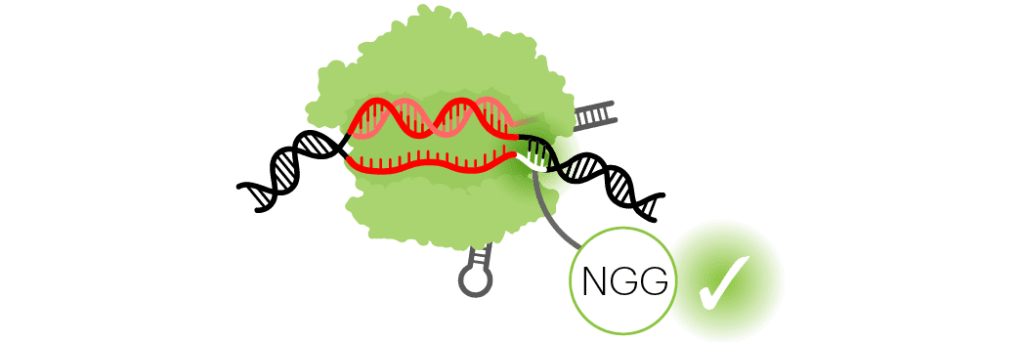
The Cas9 protein from Streptococcus pyogenes recognizes the PAM, “NGG.” The “N” means any genetic letter (A, T, G, or C). Basically, as long as there’s a “GG” one nucleotide away from the target, Cas9 will bind to it. Other Cas9s and other CRISPR systems have their own PAMs.
How is it that there’s always a PAM next to the intended target in the phage genome? The CRISPR system makes sure of this during the first step of CRISPR immunity, acquisition. When a phage injects its DNA into a bacterium, Cas1, Cas2, and another protein called Csn2 work together with Cas9 to choose appropriate pieces of the phage DNA, called prespacers, to insert into the CRISPR array. Indeed, these proteins only choose prespacers that are next to PAMs. The diligent activities of Cas1, Cas2, Csn2, and the PAM requirement enable bacteria to defend themselves from phages while avoiding having their own DNA cleaved.
🎥 | For an in-depth overview of the PAM and its importance in both natural CRISPR immunity and in genome editing applications, watch our whiteboard video below.
CRISPR diversity
CRISPR systems all work a little bit differently
HOW ARE CRISPR SYSTEMS DIFFERENT FROM ONE ANOTHER?
VOCABULARY
Hairpin/stem-loop, nuclease, effector
Bacteria have evolved a wide variety of CRISPR systems to defend themselves. At its simplest, a CRISPR-Cas system is composed of a set of cas genes and a CRISPR array. Microbes can have different combinations of cas genes, and each unique set is defined as a different CRISPR system. The Cas proteins that make up the system are like a team, each with unique roles that work together to accomplish a goal. Imagine a sports league — everyone’s playing the same game, but not every team is the same.
All CRISPR systems contain the cas1 gene. cas1 encodes the Cas1 protein, which adds new spacers to the CRISPR arrays so bacteria can fend off new phages. A CRISPR array can have anywhere between one spacer and hundreds of spacers.
What varies between systems? For just about every aspect of CRISPR immunity, nature has come up with a few variations. Let’s consider each step of CRISPR immunity, one by one.
DIFFERENCES IN ACQUISITION, CRISPR RNA BIOGENESIS, AND SURVEILLANCE COMPLEXES
- Acquisition – This step tends to be pretty similar across systems, but Cas1 and Cas2 get help from a variety of additional proteins that vary by system, including other Cas proteins and sometimes host proteins that aren’t encoded next to the cas genes and play other roles in the cell.
- crRNA biogenesis – Various Cas proteins cut pre-crRNAs into mature crRNAs. Sometimes there are dedicated Cas proteins for this job. Sometimes the same protein that accomplishes the interference step also processes its crRNA. Other times, unrelated bacterial proteins participate too. The make-up of the processed crRNA guides can vary between systems as well. crRNAs are typically a single strand of RNA that folds into a hairpin or stem-loop structure at one end. However, the presence, size, shape, and location of the hairpin within the RNA vary. Sometimes crRNAs also need additional RNAs to fully function. For example, the common CRISPR-Cas9 system uses both a crRNA and an additional piece of RNA called a tracrRNA.
- Interference – This step features the most variation between systems and serves as the basis for their classification. CRISPR-Cas systems are divided into two main classes. Class 1 systems have a large, multi-protein surveillance complex that finds nucleic acid targets. Sometimes that complex cuts the target directly, and sometimes it activates a separate cutting protein, called a nuclease, or multiple separate nucleases, for destruction. Class 2 systems, on the other hand, use a single protein to find and cleave targets. Each of these classes is further divided into types and subtypes that vary based on which set of casgenes they use.While we mostly discuss the DNA-targeting Cas9 system in CRISPRpedia, some Cas enzymes cleave RNA, and some systems destroy both DNA and RNA. Different systems usually recognize unique PAMs.
Perhaps the greatest variability comes from how CRISPR systems destroy nucleic acids. The Cas3 enzyme, found in type I systems, moves along target DNA, chopping it up like a lawn mower. Some surveillance complexes cut the same target in multiple places. Other CRISPR enzymes, like Cas9, cut just once. Finally, some CRISPR systems don’t limit cutting to their matching target but instead, once a target has been identified, begin cutting up any nearby pieces of DNA or RNA too.
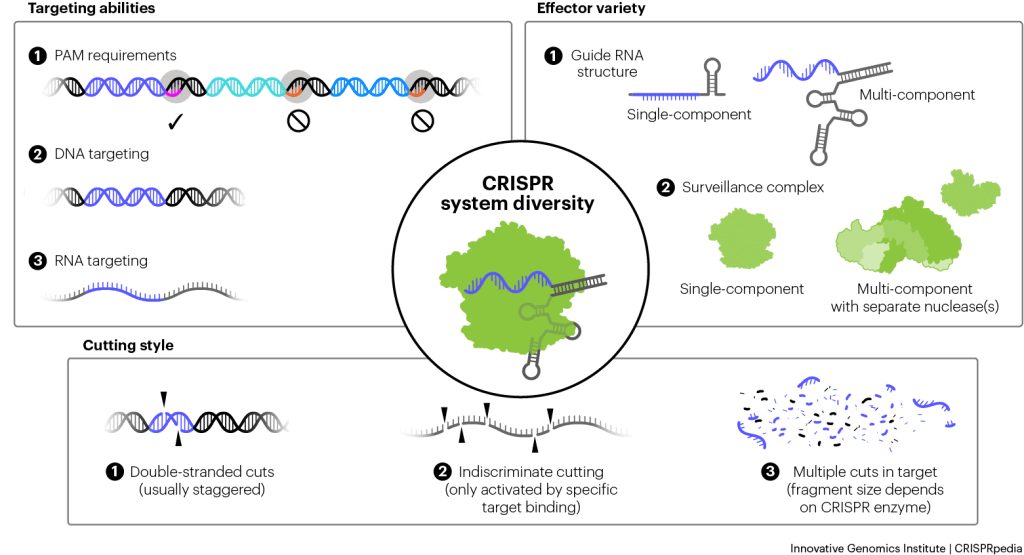
As scientists continue studying CRISPR immunity and unearthing new systems, more and more variations are revealed.
CRISPR SYSTEM VARIETY LETS BACTERIA FEND OFF DIVERSE THREATS
The number of CRISPR systems can be overwhelming and new ones are still being discovered. Why did so many variations evolve?
Different CRISPR-Cas systems allow bacteria to respond to different types of phages. For example, while most systems target DNA, some can cut RNA, and still others can cut both RNA and DNA. This comes in handy since some phages have RNA genomes, and even those with DNA genomes need to make RNA during the course of infection. Some Cas1–Cas2 complexes can even integrate RNA as new spacers.
Beyond responding to different phages, it can be helpful for bacteria to have multiple CRISPR systems as “back-up” in case one fails.
We encourage you to use and adapt CRISPRpedia illustrations for non-commercial, educational purposes. Tweet us @igisci to show us how you’re using them! ALL uses and modifications must follow the Creative Commons BY-NC-SA 4.0 International License.
SCIENTIFIC REVIEWER
Jennifer Doudna is a Professor of Molecular and Cell Biology and Chemistry at the University of California, Berkeley, and recipient of the 2020 Nobel Prize in Chemistry. The Doudna lab studies the fundamental biological processes involving RNA molecules.
Jennifer is also the Executive Director of the Li Ka Shing Center for Genomic Engineering, the Li Ka Shing Chancellor’s Chair in Biomedical and Health Sciences, and a member of the Howard Hughes Medical Institute, Lawrence Berkeley National Lab, the National Academy of Sciences, and the American Academy of Arts and Sciences.
HOW TO CITE
Doudna, J.A. (2022) CRISPR in Nature. In M.L. Hochstrasser et al. (Eds.) CRISPRpedia. Berkeley: Innovative Genomics Institute, University of California, Berkeley. Retrieved from: https://innovativegenomics.org/crisprpedia/crispr-in-nature/ (Last updated: September 12, 2022.)
SHARE PAGE