Like the Borg of Star Trek, These ‘Aliens’ Assimilate DNA from Other Microbes
Only a meter or two below our feet dwells a wealth of microbes whose riches remain largely unexplored. It’s a realm where bacteria, bacteria-like organisms called archaea and fungi mingle with viruses and other non-living bits of DNA or DNA — all living with, in or on one another.
In that alien world, researchers have now found large DNA molecules that aren’t quite viruses, which are DNA or RNA wrapped in proteins, but that seem to have infected archaea and acquired along the way a slew of genes from their archaeal hosts.
The researchers dubbed these microbes Borgs because, analogous to the fictional Borg aliens from the world of Star Trek, they assimilate pieces of the microbes they infect.
The fact that real-life Borgs contain repetitive DNA sequences between genes and even within genes, analogous to the Clustered Regularly Interspaced Short Palindromic Repeats in bacteria that gave CRISPR its name, makes Jill Banfield of the University of California, Berkeley, suspect there may be applications of Borgs that are just as revolutionary. Banfield, a UC Berkeley professor of earth and planetary science and of environmental science policy and management and a faculty scientist at Lawrence Berkeley National Laboratory (Berkeley Lab), is the senior author of a paper about Borgs that appears in this week’s edition of the journal Nature.
“Imagine a strange foreign entity, neither alive nor dead, that assimilates and shares important genes. … A floating toolbox, likely full of blueprints, some that we may one day harness, like CRISPR,” Banfield tweeted last year after she uploaded a preprint of the paper to bioRxiv.
Banfield first brought the bacterial CRISPR system to the attention of UC Berkeley colleague Jennifer Doudna in 2006. Doudna, UC Berkeley professor of chemistry and of molecular and cell biology and an expert on RNA, subsequently teamed up with French researcher Emmanuelle Charpentier to explore how the CRISPR system and an enzyme called Cas9 work in bacteria and turned the enzyme into a powerful research tool that earned Doudna and Charpentier the 2020 Nobel Prize in Chemistry.
While Borgs are too new to know exactly what they are and what secrets they may reveal, Banfield is optimistic.
“This discovery started in deep mud and was brought to light by an analysis of around 10 billion DNA snippets,” she wrote in her 2021 Twitter thread. “That such an approach could reveal something with potentially global ramifications! … In 2021, I will again sit across the table from Jennifer Doudna (@doudnalab) and we will talk about how we might begin to explore the technological and environmental importance of this discovery.”
In a Nature commentary about the paper, Christian Rinke of the University of Queensland in Australia wrote, “It will be exciting to see whether Borgs affect the carbon cycle in their native habitats, in particular regarding the consumption of methane. It may be that, in contrast to those in Star Trek, the Borgs in our Universe use assimilation to save rather than to conquer a world.”
Some archaea generate methane; others eat it up
In the Nature paper, Banfield, doctoral candidate Basem Al-Shayeb, postdoctoral fellow Marie Schoelmerich and their colleagues describe what they’ve learned about Borgs from sequencing Borg DNA and the DNA of their archaeal hosts.
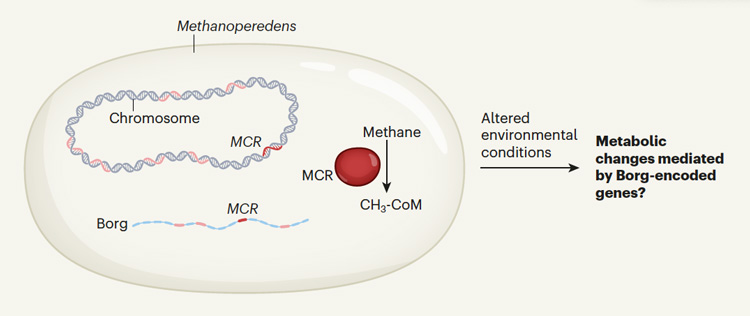
All of the 19 Borg genomes they include in the paper contain genes that seem to enhance the ability of their host archaea to harvest resources in the soil, such as to metabolize methane or fix nitrogen. This may allow the host to modulate its metabolic capacity under various environmental conditions, revving up the archaea’s metabolism when methane or nitrogen is particularly abundant.

Methane is a common ingredient in wetland soils, produced by methanogenic archaea when oxygen is absent, that is, in anaerobic conditions. But some bacteria also consume methane when oxygen is available, as do certain archaea when oxygen is not available. The methane-consuming archaea that host Borgs reverse the process used by methane-producing archaea in order to eat methane and turn it into the complex carbohydrates they need to survive. The genes for methane oxidation found in Borgs appear to have been acquired from their archaeal hosts, which are all from the family Methanoperedens.
The researchers suspect that Borgs may play an outsized role in the subsoil, revving up the removal of methane and fixation of nitrogen by their host Methanoperedens. If so, enhancing the population of Borgs inside their archaeal methane-eating hosts could help remove methane — a greenhouse 30 times more potent than carbon dioxide — from the atmosphere.
“Right now, microbes are the methane sink of the planet,” said Al-Shayeb. “They’re very prevalent, for example, in rice paddies and in farmlands and in these anaerobic, very low-oxygen environments, and they contribute to the removal of methane from the environment. The possible applications in agriculture or in methane reduction are very exciting — maybe one day it will pave the way to make a dent in climate change.”
At the moment, no one has been able to grow Borg-containing Methanoperedens archaea in the lab. But researchers in the Innovative Genomics Institute (IGI), which Doudna co-founded and is now the leading center of CRISPR research in the world, are focused on changing that.
In a 2021 IGI video, postdoc Marie Schoelmerich and doctoral candidate Basem Al-Shayeb discussed their discovery of Borgs — mysterious DNA elements that reside in microorganisms and which contain genes needed for methane breakdown.
“We’re setting up bioreactors that contain soil cores from the site where we’ve been able to find Borgs. And then we are going to incubate these soils with methane, and we’re going to attempt getting a stable community and ideally enriching for Borgs and Methanoperedens in the lab, to follow up on our hypotheses that they are augmenting methane oxidation rates,” Schoelmerich said.
The researchers discovered Borgs after sequencing the genomes of microbial communities in various subsurface environments, including vernal pools in Northern California and soil from a remediation site in Rifle, Colorado. They’ve since found them in many oxygen-poor environments they’ve sampled.
The Borg DNA stood out as unique from the typical genomes of bacteria and archaea and the other life forms that dwell among them: viruses, which are typically short lengths of DNA or RNA encapsulated by protein, and plasmids, which are short lengths of DNA that form rings and live inside most bacteria.
“We were scratching our heads for a long time trying to determine whether this is a plasmid, whether it’s a virus,” Al-Shayeb said. “Through a process of elimination, we decided it didn’t fit into either bucket.”
For one thing, Borg genomes were huge, ranging up to 1.4 million base pairs in length — one-third the length of the entire Methanoperedens genome. Compare this to the genomes of viruses, which are one-tenth as big, or circular plasmids, which are typically tens of thousands of base pairs in length. And each Methanoperedens cell can contain many separate Borgs — sometimes more than eight, Schoelmerich said — which means that the combined Borg genomes far exceed the size of the genome of their host.
Borgs also contain a lot of the same DNA as in their host archaea.
“We found that there were more genes shared between the Borgs and the host Methanoperedens than there were genes shared between the different Borgs or any other sort of extra chromosomal genetic element that was previously described,” Al-Shayeb said. “And the genes that were shared with Methanoperedens included a lot of metabolism genes: genes involved with nitrogen fixation, with methane oxidation, even extracellular electron transfer.”
And each Borg encodes a unique set of proteins, perhaps picked up from various archaeal hosts over time.
“There is evidence that different types of Borgs sometimes coexist in the same host Methanopreredens cell. This opens the possibility that Borgs could be spreading genes across lineages,” Banfield said.
It’s still unclear why or how Borgs assimilated these genes and what effect — positive or negative — Borgs have on their host. But Schoelmerich said that the total genome size of all Borgs within a cell would be a burden, and thus would likely offer some benefit to the host.
More Borg genomes
Banfield and her team posted a preprint detailing an additional six Borg genomes on bioRxiv in May, when Banfied tweeted: “Today we unveil 6 new polished genomes for Borgs, enigmatic non-living entities found in the microscopic universe of dark, cold, wet soil.”
Schoelmerich, first author of the preprint, focuses mostly on the repetitive sequences of DNA in Borgs, which resemble tandem repeats in CRISPR, and in Borgs seem to evolve quickly, generating disordered regions in proteins — like randomly assembled Legos lacking any coherent structure — that may help Borgs adapt quickly to a changing environment within their host cell.
“We think the Borg genome evolves rapidly to fine-tune its association with the specific host,” Schoelmerich said. “These tandem repeat regions can be a mechanism by which this specific association is ensured.”
Much work remains to determine exactly what role Borgs play in the soil environment, as well as within the Methanoperedens archaea, and whether they can be tweaked to address climate change.
“It’s possible that we can enrich Borg-containing Methanoperedens in soils naturally and change that thermodynamic equilibrium that’s now on the side of methane production and push that back so it’s more on the side of methane consumption,” Schoelmerich said. “If we can understand more about the metabolism and how they’re actually able to oxidize methane, and which other elements such as Borgs play a role in this, then maybe we can reverse the methane emissions. That would be amazing. We’re excited about it.”
Other co-authors of the Nature paper are Doudna, Jacob West-Roberts, Luis Valentin-Alvarado, Rohan Sachdeva, Susan Mullen and Alexander Crits-Christoph of UC Berkeley; Michael Wilkins of Colorado State University; and Kenneth Williams of Berkeley Lab. The research was supported by the U.S. Department of Energy (DEAC02-05CH11231), the Chan Zuckerberg Biohub and the Innovative Genomics Institute.
Media Contact:
Andy Murdock, andymurdock@berkeley.edu
 By
Robert Sanders
By
Robert Sanders



