

The Banfield lab investigates the role of soil microbes in greenhouse gas emissions
Jill Banfield has collected soil and water samples from acid mines, hot springs, manure ponds, bioreactors, and more. But these days, you might find her somewhere a little more pastoral: knee-deep in a rice paddy. Banfield and her team are studying the microorganisms that live in rice paddy soils, with the ultimate aim of understanding how they contribute to greenhouse gas emissions — and how to intervene.
Rice Cooks You
Greenhouse gas emissions are the main cause of climate change, and agriculture is one of the biggest sources of greenhouse gas emissions. While there is a lot of focus on industrial animal agriculture and emissions, rice cultivation is actually one of the largest sources.
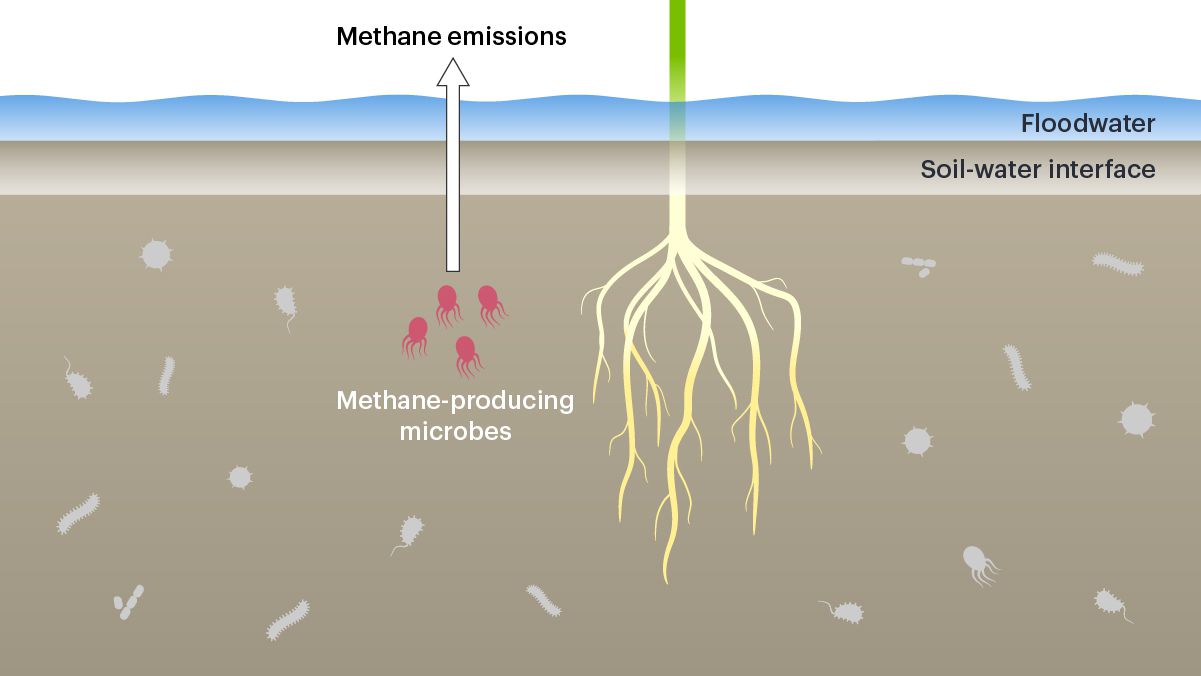
Rice is traditionally grown in flooded paddies, where there are constantly a few inches of water above the soil. In these conditions, certain microbes in the soil make energy in ways that produce methane — a greenhouse gas many times more potent than carbon dioxide. In a 20-year period, atmospheric methane has a global warming potential of more than 80 times greater than carbon dioxide. In short: methane is bad for the climate, and rice farming makes a lot of it.
Under the Surface
Scientists know that bacteria and other unicellular organisms make methane, but how it’s produced or what controls the makeup of the microbial community below the surface in rice paddies remains poorly understood. Postdoctoral researcher Bethany Kolody, graduate student Jack Kim, and others in the Banfield lab are using a variety of methods, including metagenomics, geochemistry, and machine learning to learn about the members of these soil microbial communities, how different microbes interact with each other and with rice plants to make and release methane, and how we might be able to reduce methane emissions.
“It’s amazing that nobody studied the rice soil microbiome over the growing season with genomic methods, given the role microbes play in producing — and consuming — methane,” says Banfield.
Banfield and her lab pioneered genome-resolved metagenomics, an approach that allows identification of unknown microbes growing in the wild in mixed populations, and, based on their genomes, find out things like how they make energy, protect themselves from predators, or whether they produce or consume methane.
A recent gift of $3 million to IGI climate projects is providing funding for this crucial work and other projects that are expanding IGI’s research into nature-based climate change solutions enabled by genomic technologies. What Banfield and her team learn about rice paddy microbes and plant-microbe interactions will determine the next steps and what a scalable intervention could look like.
We Are the Borg
Banfield’s team recently made a related discovery — mysterious DNA elements known as Borgs, which appear to reside in methane-eating microbes. The Borg DNA, named for its ability to assimilate genes from other microbes, contains additional copies of genes needed for methane breakdown. While there’s much more to learn about Borgs, it’s likely they have an important role in methane metabolism.
“We don’t know yet whether we’ll find them in rice paddies,” says Banfield, “But from what we know, they follow the methane.”
This project is part of a set of related projects working toward the vision of creating a net-zero farm of the future. Other projects from IGI researchers are working to minimize farmer inputs, optimize crop photosynthesis, and increase carbon sequestration in agricultural soils.
Watch this video to learn more about Borgs from IGI researchers Marie Schoelmerich and Basem Al-Shayeb.



