Genomics
Institute

Glossary
Glossary
Our illustrated glossary explains key CRISPR-related terms – in English and Spanish – without jargon. The 80+ original icons are downloadable as editable vector graphics and free to use for non-commercial purposes under a Creative Commons license. Scroll to the end of the page for details on image usage and download options.
For a more education content, check out CRISPRpedia and CRISPR Made Simple.
-
Agrobacterium
A type of soil bacterium that can transfer its own genes into plant cells and integrate into their genomes. Often used as a way to add new genes to crop plants.
-
AlleleOne of several possible versions of a gene. Each one contains a distinct variation in its DNA sequence. For example, a “deleterious allele” is a form of a gene that leads to disease.
-
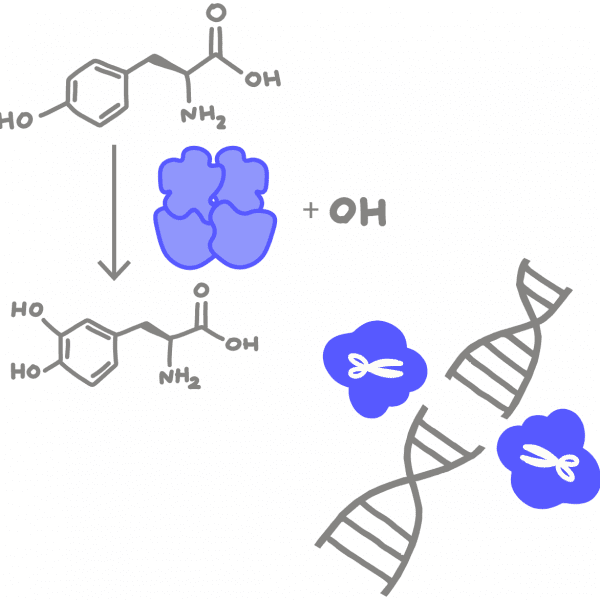
Amino acidThe chemical building block of proteins. During translation, different amino acids are strung together to form a chain that folds into a protein.
-
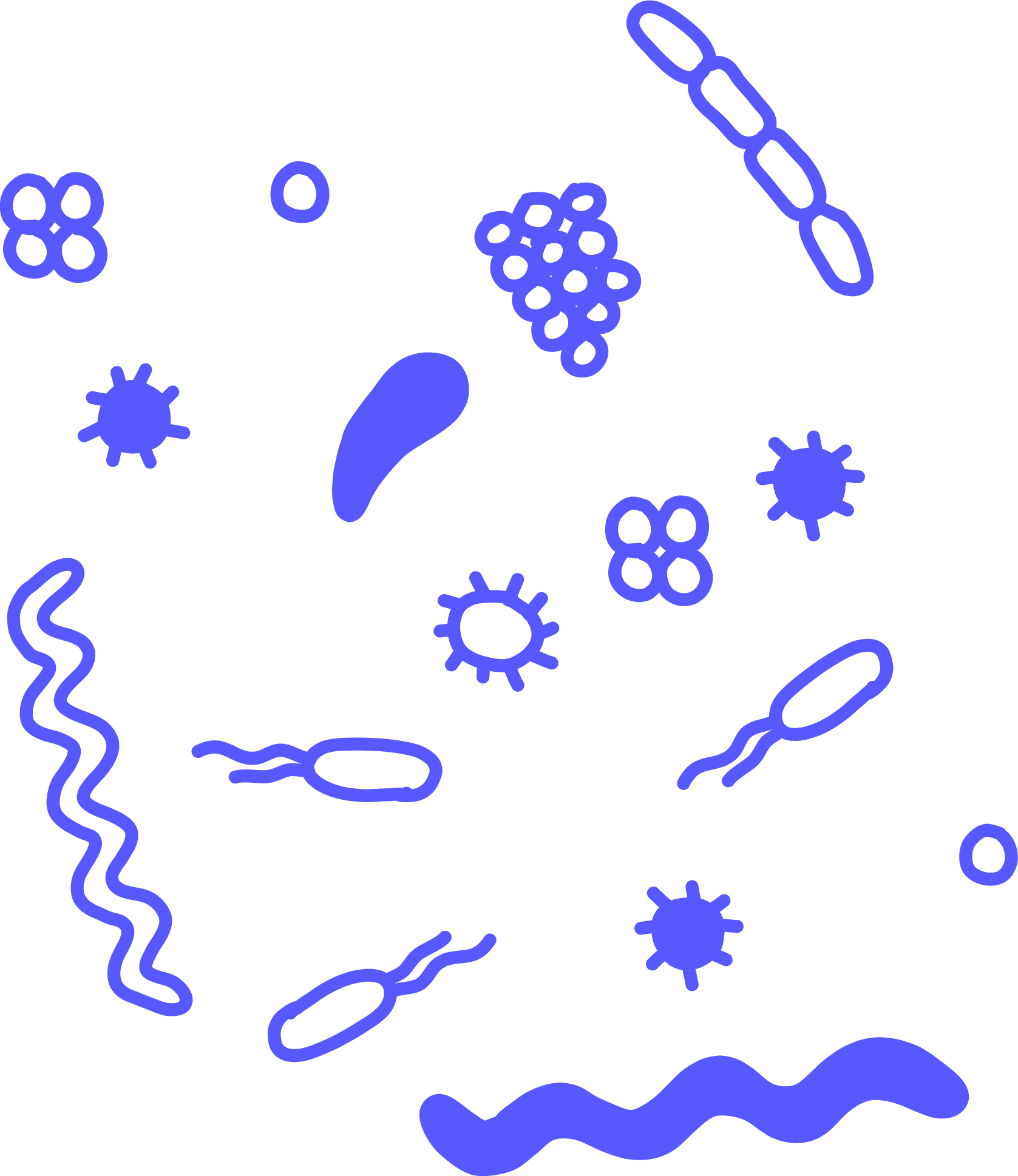
ArchaeaMicrobes that look similar to bacteria but are actually more closely related to eukaryotes, such as humans. Archaea are single-celled organisms that don’t have a nucleus and can only be seen with a microscope. They’re found in many different habitats, and many of the first known examples were found in extreme environments.
-
BacteriaAn abundant type of microbe. These single-celled organisms are invisible to the naked eye, don’t have a nucleus, and can have many shapes. They’re found in all types of environments, from Arctic soil to inside the human body. Most bacteria are not harmful to human health, but certain pathogenic bacteria can cause illness.
-
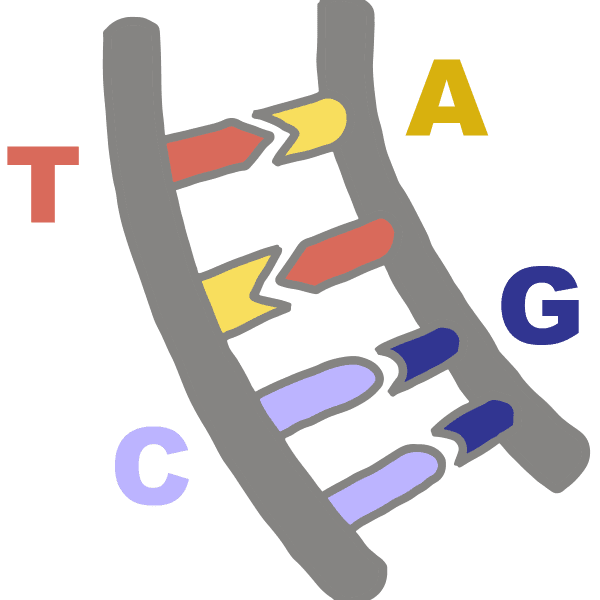
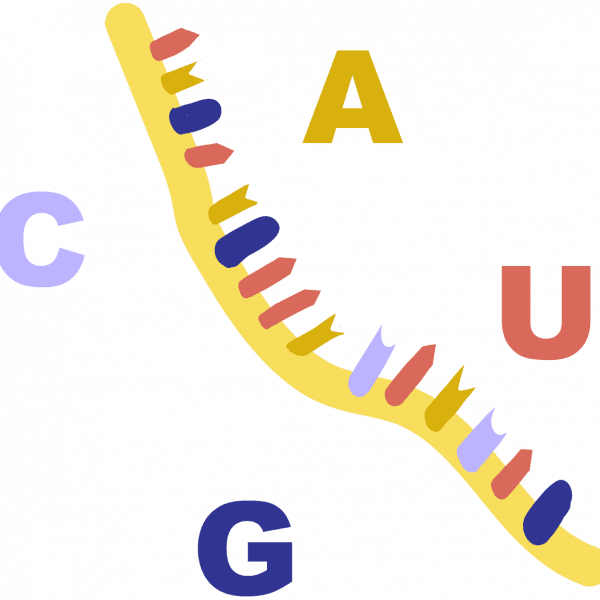
BaseThe four “letters” of the genetic code (A, C, T, and G) are chemical groups called bases or nucleobases. A = adenine, C = cytosine, T = thymine, and G = guanine. Instead of thymine, RNA contains a base called uracil (U).
-
Base editing
A type of genome editing that uses CRISPR components to directly change a limited set of single DNA letters to other letters without making breaks in the DNA. Base editors are made by fusing (attaching) DNA-modifying enzymes to DNA-targeting Cas proteins like Cas9.
-
Base pairDifferent chemicals known as bases or nucleobases are found on each strand of DNA. Each base has a chemical attraction for a particular partner base, known as its complement. C matches up with G, while A pairs with T or U. These bonded genetic letters are called base pairs. Two strands of DNA can zip together to form a double-helix shape when complementary bases match up to form base pairs.
-
CancerA type of disease caused by uncontrolled growth of cells. Cancerous cells may form clumps or masses known as tumors, and can spread to other parts of the body through a process known as metastasis.
-
CasAbbreviation of CRISPR-associated, may refer to genes (cas) or proteins (Cas) that protect bacteria and archaea from viral infection.
-
Cas3
A protein derived from the CRISPR-Cas bacterial immune system that has been co-opted for genome engineering. Cas3 is a standalone nuclease-helicase that is recruited to targets by a separate, crRNA-guided complex called Cascade. Once Cascade identifies a complementary DNA target, it recruits Cas3 to move along the DNA, cutting as it goes. In CRISPR immunity, cutting phage DNA prevents phages from replicating and destroying the host cell. When used for genetic engineering, this approach can be used to delete thousands of base pairs around the target site.
-
Cas9A protein derived from the CRISPR-Cas bacterial immune system that has been co-opted for genome engineering. Uses an RNA molecule as a guide to find a complementary DNA sequence. Once the target DNA is identified, Cas9 cuts both strands. Has been compared to “molecular scissors” or a “genetic scalpel.” In CRISPR immunity, cutting viral DNA prevents it from destroying the host cell. In genome engineering, cutting genomic DNA initiates a repair process that ends up making a change or “edit” to its sequence.
-
Cas12
A protein derived from the CRISPR-Cas bacterial immune system that has been co-opted for genome engineering. Uses an RNA molecule as a guide to find a complementary DNA sequence. Once the target DNA is identified, Cas12 cuts both strands. Has been compared to “molecular scissors” or a “genetic scalpel.” In CRISPR immunity, cutting viral DNA prevents it from destroying the host cell. In genome engineering, cutting genomic DNA initiates a repair process that ends up making a change or “edit” to its sequence.
Note that Cpf1 (Cas12a), C2c1 (Cas12b), C2c3 (Cas12c), CasY (Cas12d), CasX (Cas12e), Cas14 (Cas12f), and CasPhi (Cas12j) are all Cas12 family members. Our illustration is based on Cas12a.
-
Cas13
A protein derived from the CRISPR-Cas bacterial immune system that has been co-opted for genome engineering and CRISPR-based diagnostics. Uses an RNA molecule as a guide to find a complementary RNA sequence. Once the target RNA is identified, Cas13 cuts both the target and nearby, “collateral” RNA. In CRISPR immunity, cutting RNA like this is thought to cause “altruistic” cell suicide, preventing the spread of phages to neighboring cells. In genome engineering, an engineered version of Cas13 can be used to make a change or “edit” to targeted RNA sequences. In CRISPR-based diagnostics, Cas13’s collateral cutting activity can generate a fluorescent signal to indicate the presence of a pathogenic target RNA.
-
Cascade
A protein complex derived from the CRISPR-Cas bacterial immune system that has been co-opted for genome engineering. Cascade is composed of multiple Cas proteins. The complex uses a crRNA as a guide to find a complementary DNA sequence. Once the target DNA is identified, Cascade recruits a separate nuclease-helicase called Cas3 to move along the DNA, cutting as it goes. In CRISPR immunity, cutting phage DNA prevents phages from replicating and it from destroying the host cell. When used for genetic engineering, this approach can be used to delete thousands of base pairs around the target site.
-
CellThe basic unit of life. The number of cells in a living organism ranges from one (e.g. yeast) to quadrillions (e.g. blue whale). A cell is composed of four key macromolecules that allow it to function (protein, lipids, carbohydrates, and nucleic acids). Among other things, cells can build and break down molecules, move, grow, divide, and die.
-
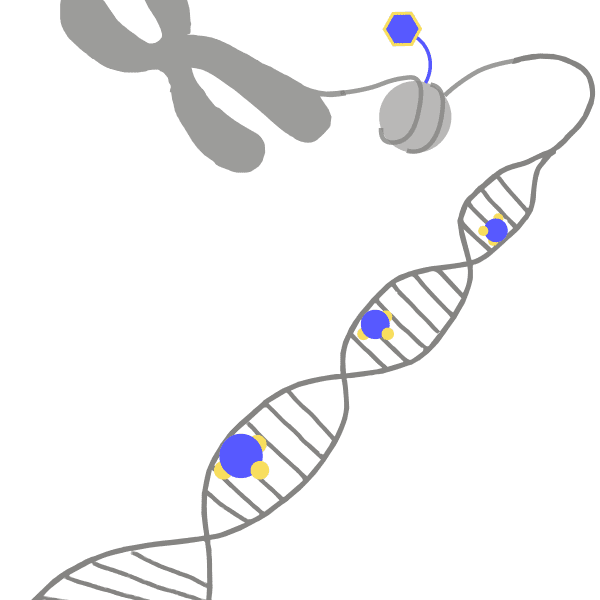
ChromosomeThe compact structure into which a cell's DNA is organized, held together by proteins. The genomes of different organisms are arranged into varying numbers of chromosomes, and human cells have 23 pairs.
-
CleaveThe scientific term for cut or break apart. Typically refers to splitting apart a long polymeric molecule like DNA, RNA, or protein. For example, a nuclease like Cas9 can be directed to cleave DNA at a specific location.
-
ComplementaryDescribes any two DNA or RNA sequences that can form a series of base pairs with each other. Each base forms a bond with a complementary partner. T (DNA) and U (RNA) bond with A, and C complements G. For example, in CRISPR immunity, the spacer sequence in a guide RNA is complementary to a sequence found in a viral genome. When the RNA bases pair with complementary DNA bases from an invading virus, the Cas9 protein will cut the target to stop the viral infection.
-
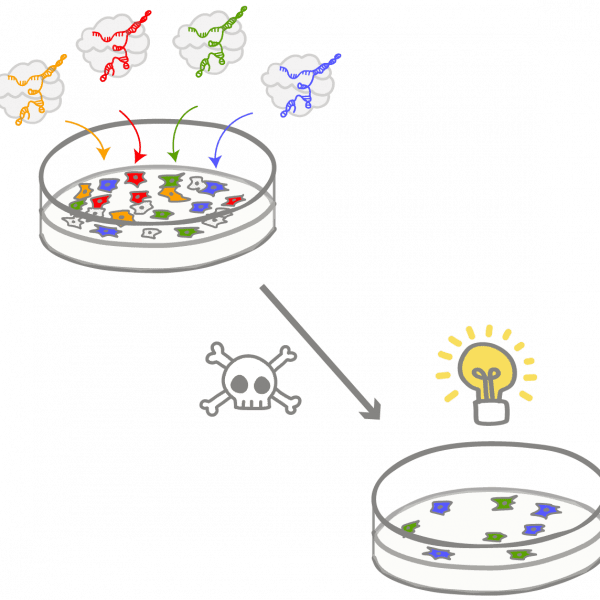
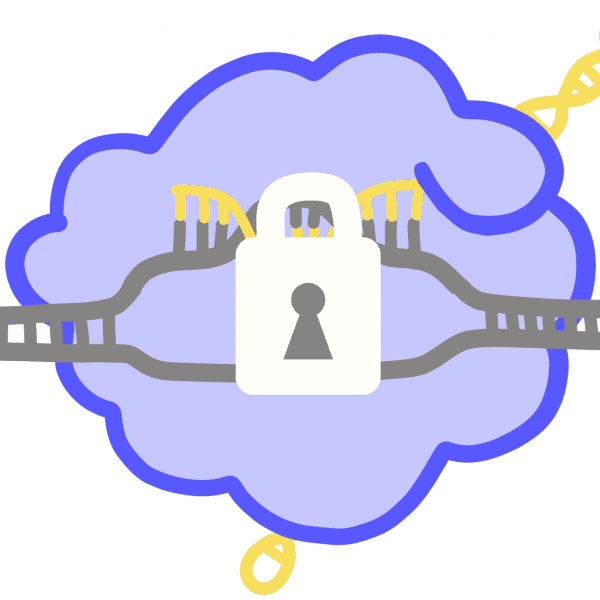
CRISPRPronounced “crisper.” An adaptive immune system found in bacteria and archaea, co-opted as a genome engineering tool. Acronym of “clustered regularly interspaced short palindromic repeats,” which refers to a section of the host genome containing alternating repetitive sequences and unique snippets of foreign DNA. CRISPR-associated surveillance proteins use these unique sequences as molecular mugshots as they seek out and destroy viral DNA to protect the cell.
-
CRISPR RNA (crRNA)During CRISPR immunity, the host cell generates crRNA molecules, each containing one spacer that is complementary to a portion of a viral genome. crRNAs guide CRISPR immune proteins to find and destroy matching invader sequences.
-
CRISPR screeningA technique that lets scientists see the effects of turning gene expression up or down with CRISPRa and CRISPRi. Instead of checking one gene at a time, a single CRISPR screen can provide information about thousands of different genes at a time.
-
CRISPRa and CRISPRiCRISPRa stands for CRISPR activation and CRISPRi stands for CRISPR interference or inhibition. Both are methods for fine-tuning gene expression. If a gene were a car, CRISPRa is the gas pedal and CRISPRi is the brake. Using CRISPRa to activate a gene increases protein production. Using CRISPRi to “turn down” a gene reduces the number of protein products made from that gene.
-
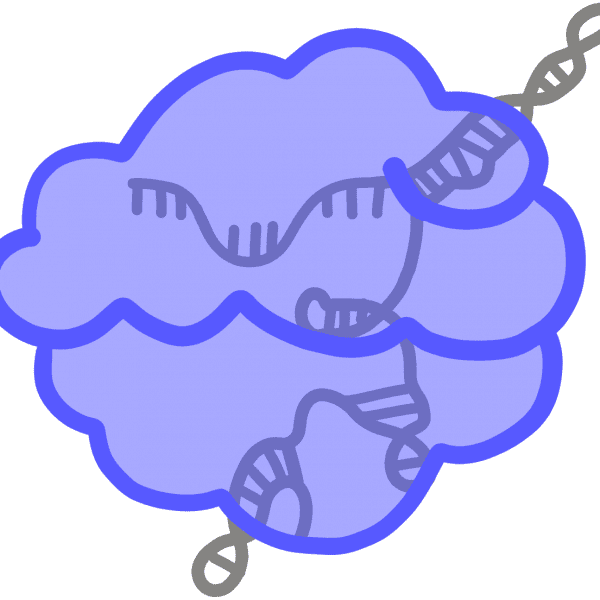
dCas9Catalytically inactive, or “dead,” Cas9. This mutated version of the Cas9 protein cannot cut, but still binds tightly to a particular DNA sequence specified by the guide RNA. Can be used to physically block the process of transcription, turning off a specific gene, or to shuttle other proteins to a particular site in the genome.
-
DNAAbbreviation of deoxyribonucleic acid, a long molecule that encodes the information needed for a cell to function or a virus to replicate. Forms a double-helix shape that resembles a twisted ladder. Different chemicals called bases, abbreviated as A, C, T, and G, are found on each side of the ladder, or strand. The bases have an attraction for each other, making A stick to T while C sticks to G. These rungs of the ladder are called base pairs. The sequence of these letters is called the genetic code.
-
Double-strand break (DSB)When both strands of DNA are broken, two free ends are created. May be made intentionally by a tool such as Cas9. Cells repair their DNA to prevent cell death, sometimes changing the DNA sequence at the site of the break. Initiating or controlling this process with the intent to alter a DNA sequence is known as genome engineering.
-
Envelope delivery vehicle (EDV)
The exterior envelope of a virus that is engineered to deliver genes or genome-editing molecules without causing infection or disease. The viral genome is not present in EDVs.
-
EnzymeA molecule, typically a protein, that causes or catalyzes a chemical change. Usually an enzyme’s name describes a molecule involved in the activity it performs and ends with the suffix -ase. For example, lactase is a well-known enzyme that breaks down lactose, a sugar found in milk. Cas9 is a nuclease, an enzyme that breaks apart the backbone of nucleic acids (RNA or DNA).
-
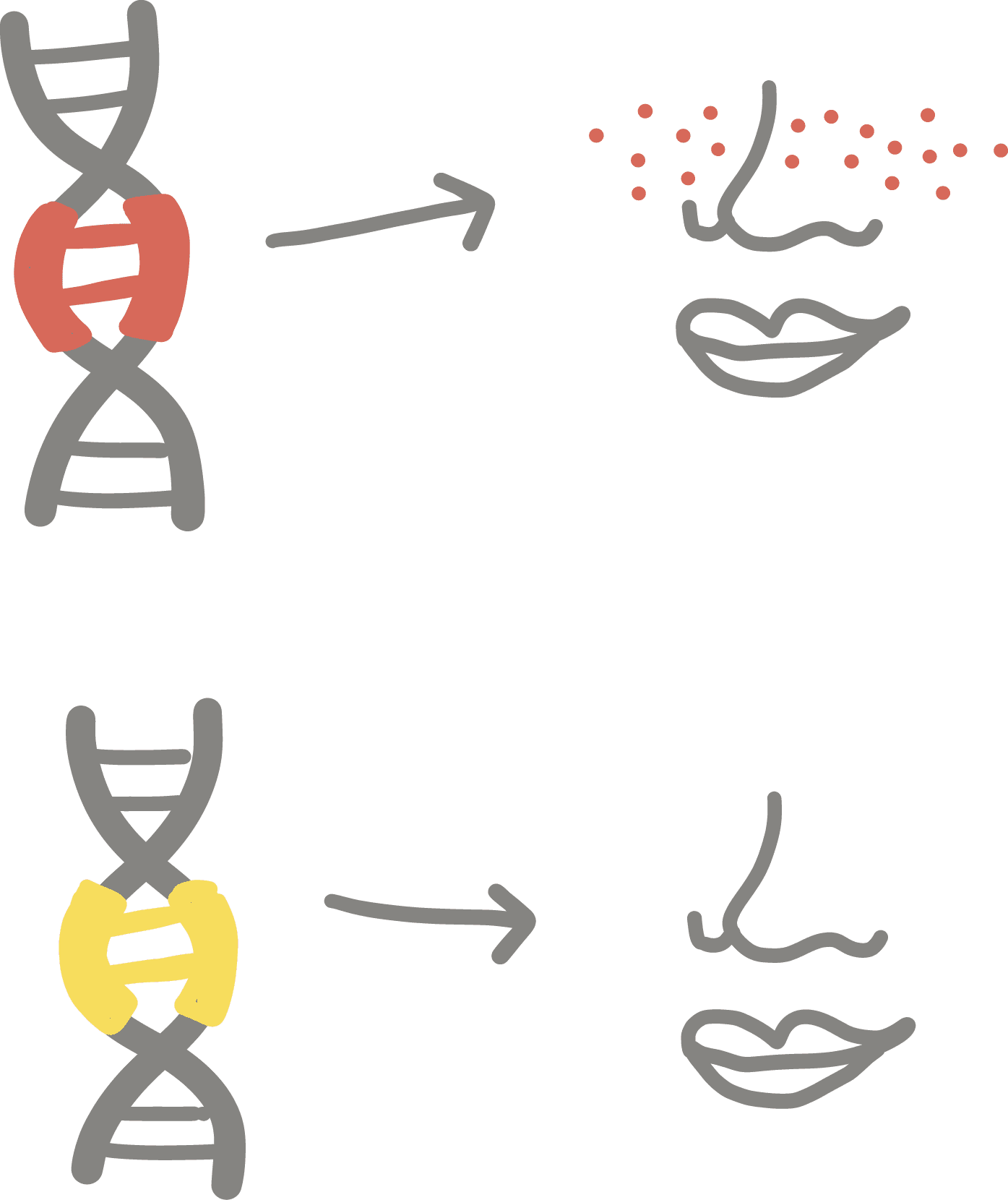
Epigenetic
Refers to changes to a cell’s gene expression that do not involve altering its DNA code. Instead, the DNA and proteins that hold onto DNA are “tagged” with removable chemical signals. Epigenetic marks tell other proteins how to read the DNA, which parts to ignore, and which parts to transcribe into RNA.
Comparable to sticking a note that says “SKIP” onto a page of a book—a reader will ignore this page, but the book itself has not been changed.
-
EukaryoteA domain of organisms whose cells contain a nucleus and other organelles. Eukaryotes are often large and multicellular (e.g. elephants) but can also exist as microscopic, single cells (e.g. yeast). This category of life includes humans. Compare to prokaryotes (bacteria and archaea).
-
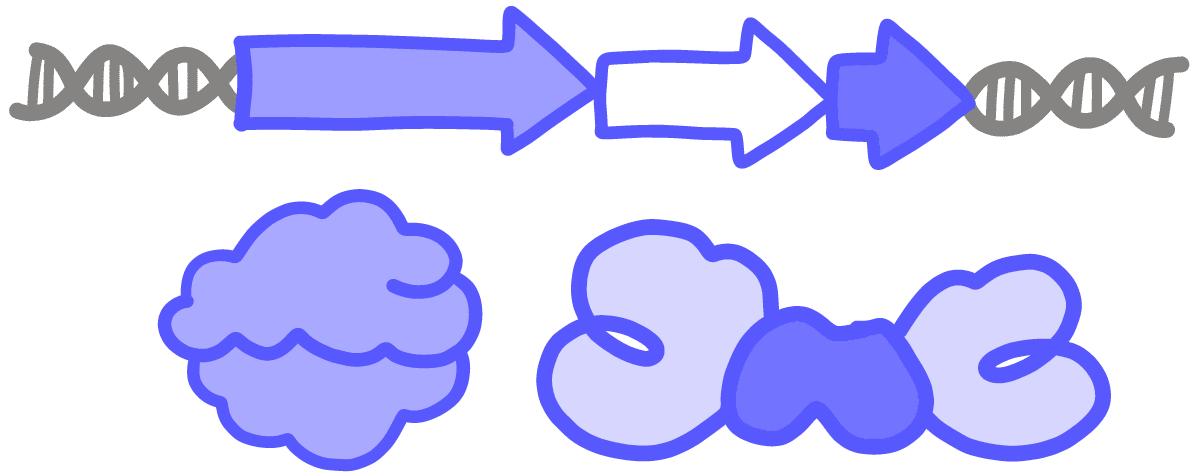
ExpressionA product being made from a gene; can refer to either RNA or protein. When a gene is turned on, cellular machines “express” this by transcribing the DNA into RNA and/or translating the RNA into a chain of amino acids. For example, a “highly expressed” gene will have many RNA copies produced, and its protein product is likely to be abundant in the cell. CRISPRi and CRISPRa are methods for turning gene expression down or up, respectively.
-
GeneA segment of DNA that encodes the information used to make a protein. Each gene is a set of instructions for making a particular molecular machine that helps a cell, organism, or virus function.
-
Gene driveA mechanism for preferential inheritance of a particular DNA sequence. Usually, offspring have a semi-random chance of inheriting a given stretch of DNA from either parent. In a scientist-designed gene drive, a gene is engineered to have a 100% chance of being passed on. Gene drives can force the inheritance of a desirable trait through a population of organisms. For example, this approach could potentially make all mosquitoes incapable of transmitting the malaria parasite.
-
Gene therapy
Delivering corrective DNA to human cells as a medical treatment. Certain diseases can be treated or even cured by adding a healthy DNA sequence into the genomes of particular cells. Scientists and doctors typically use a virus to shuttle genes into targeted cells or tissues, where the DNA is incorporated somewhere within the cells’ existing DNA. CRISPR genome editing is sometimes referred to as a gene therapy technique.
-
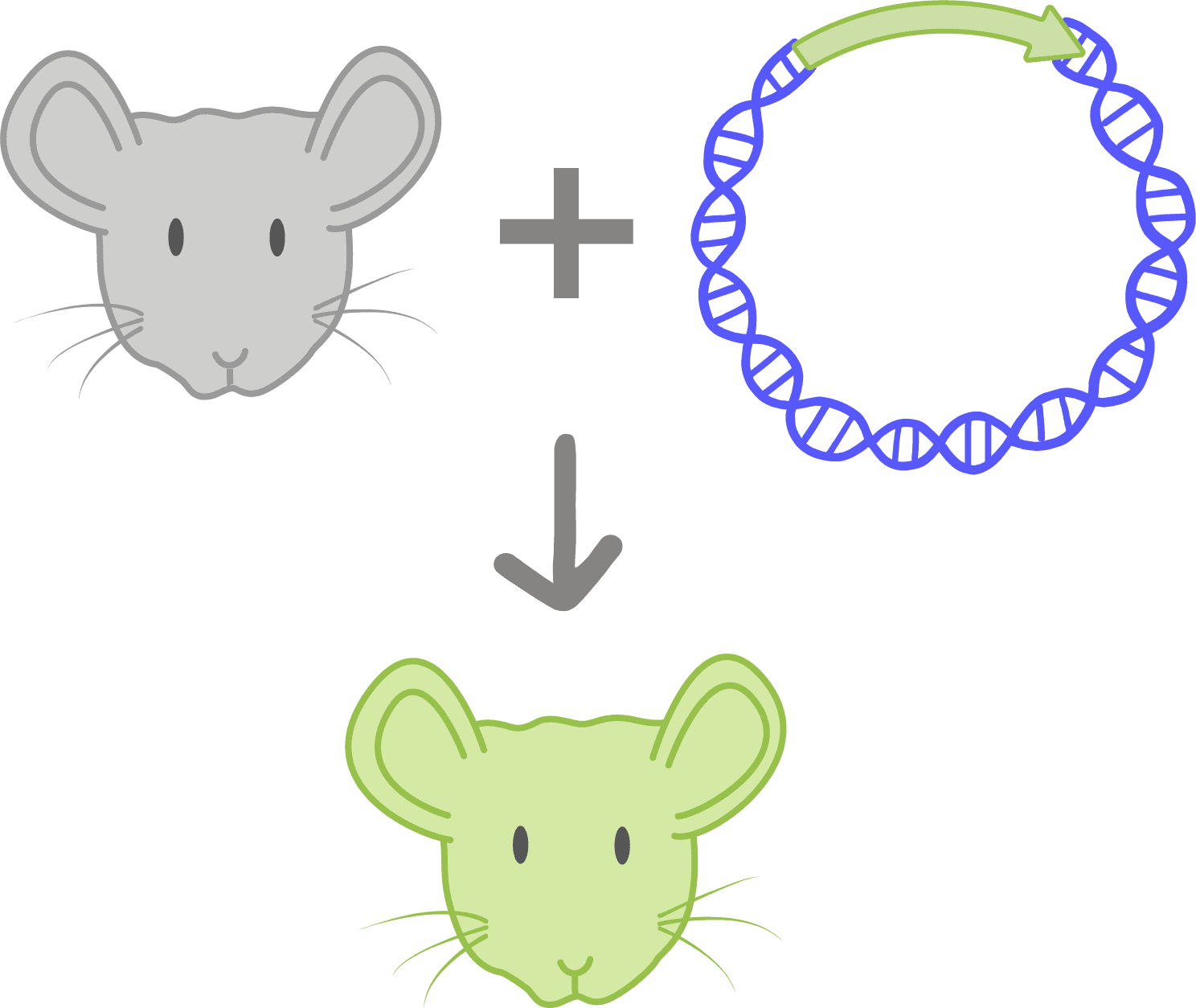
Genetically modified organism (GMO)A genetically modified organism has had its DNA intentionally altered using scientific tools. Any organism can be engineered in this manner, including microbes, plants, and animals.
-
GenomeThe entire DNA sequence of an organism or virus. The genome is essentially a huge set of instructions for making individual parts of a cell and directing how everything should run.
-
Genome editingIntentionally altering the genetic code of a living organism. Can be done with ZFNs, TALENs, or CRISPR. These systems are used to create a double-strand break at a specific DNA site. When the cell repairs the break, the sequence is changed. Can be used to remove, change, or add DNA.
-
Genome surgeryRepairing harmful DNA through a one-time genome editing procedure. Unlike taking a drug that will temporarily reduce long-term symptoms, altering a patient’s genetic code with the CRISPR-Cas9 “molecular scalpel” would permanently and directly reverse the cause of a genetic disease.
-
GenomicsThe study of the genome, all the DNA from a given organism. Involves a genome’s DNA sequence, organization and control of genes, molecules that interact with DNA, and how these different components affect the growth and function of cells.
-
Germ cellsThe cells involved in sexual reproduction: eggs, sperm, and precursor cells that develop into eggs or sperm. The DNA in germ cells, including any mutations or intentional genetic edits, may be passed down to the next generation. In contrast, the genetic material in somatic cells (all the cells in the body except for germ cells) cannot be inherited by offspring. Note that genome editing in an early embryo is considered to be germline editing since any DNA changes will likely end up in all cells of the organism that is eventually born.
-
Guide RNA (gRNA)A two-piece molecule that Cas9 binds and uses to identify a complementary DNA sequence. Composed of the CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA). Cas9 uses the tracrRNA portion of the guide as a handle, while the crRNA spacer sequence directs the complex to a matching DNA sequence.
Scientists have also formed a version of the guide RNA that consists of a single molecule, the single-guide RNA (sgRNA).
-
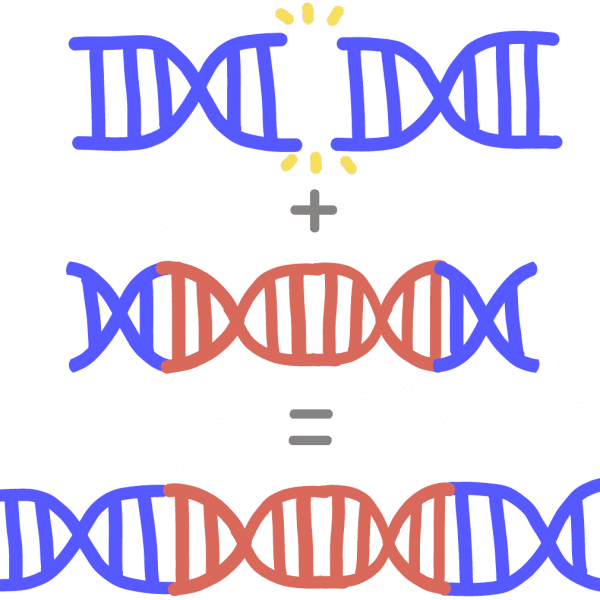
Homology-directed repair (HDR)A way for a cell to repair a break in its DNA by “patching” it with a piece of donor DNA. The donor DNA must contain similar sequences, or homology, to the broken DNA ends for it to be incorporated. HDR is a more precise repair pathway than non-homologous end joining. In genome engineering, a researcher designs and adds in the donor DNA, potentially allowing scientists to replace a disease-causing gene with a healthy copy.
-
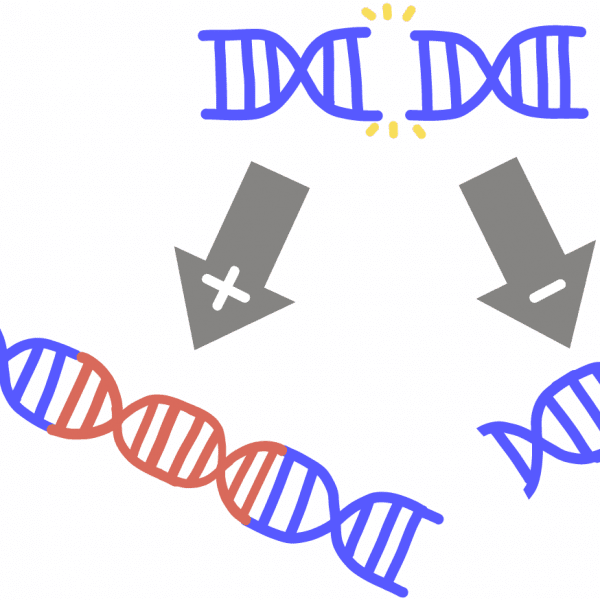
IndelAbbreviation for insertion or deletion. Refers to the random removal or addition of nucleotides from a DNA sequence. This can be enough to stop a gene from functioning (imagine removing a page from the middle of an instruction manual). Indels occur when DNA is broken and “sloppily” repaired by the cell in a process called non-homologous end joining (NHEJ).
-
Lipid nanoparticle
A drug delivery system, where therapeutic molecules are delivered inside a droplet made of lipids (fats).
-
Metagenomics
The study of a collection of genomes from a mixed community of organisms, typically microbes. Requires sophisticated computer algorithms to piece together and analyze genome sequences from the mixed sample.
-
Microbe
A microscopic organism. Can be single-celled or multicellular, and is sometimes used to refer to viruses, although they are not considered to be alive. Examples include bacteria, yeast, and algae.
-
Microbiome
A community of microorganisms in a particular environment, like soil or the human body.
-
Monogenic
Describes a trait that is determined by one gene. Compare to polygenic.
-
MutationA change from one genetic letter (nucleotide) to another. Variation in DNA sequence gives rise to the incredible diversity of species in the world, and even occurs between different organisms of the same species. While some mutations have no consequence at all, certain mutations can directly cause disease. Mutations may be caused by DNA-damaging agents such as UV light or may arise from errors that occur when DNA is copied by cellular enzymes. They can also be made deliberately via genome engineering methods.
-
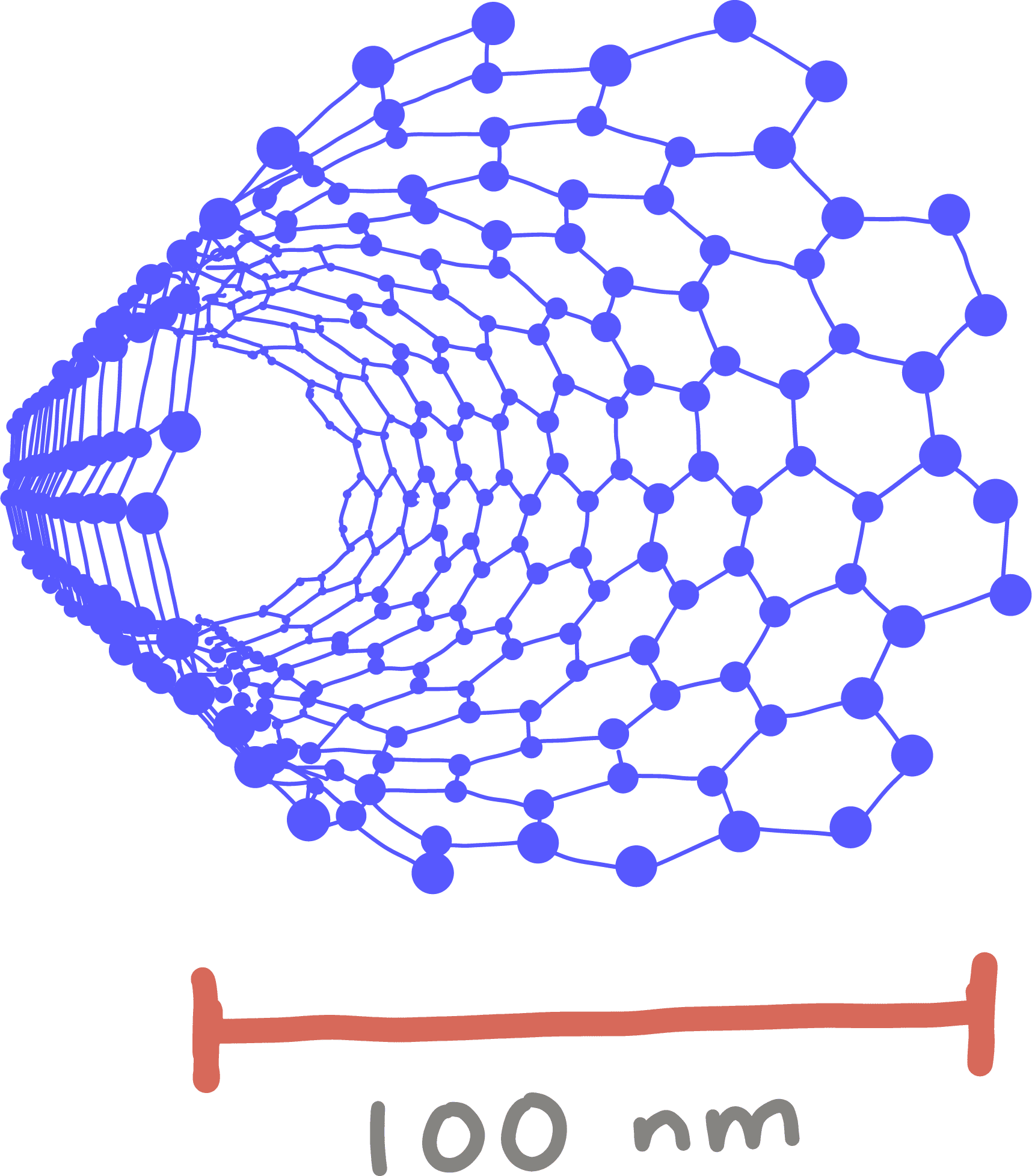
Nanomaterial
A material that is less than 100 nanometers in at least one direction – that is, less than about 1/1000 of the width of a human hair. Some materials have different properties at this small size than in their larger forms. Nanomaterials are being developed as methods to deliver genome-editing components to cells.
-
Non-homologous end joining (NHEJ)A way for a cell to repair a break in its DNA by attaching the free DNA ends. This pathway is “sloppier” than homology-directed repair, and often results in the random addition or removal of nucleotides around the site of the DNA break, causing insertions or deletions in the genetic code. In genome engineering, this allows scientists to stop a gene from working (similar to removing a page from the middle of an instruction manual).
-
NickWhen only one strand of DNA is broken, there is a gap called a nick in the backbone, but the DNA does not separate. A tool like CRISPR-Cas9 may be used to generate a nick.
-
NucleaseAn enzyme that breaks apart the backbone of RNA or DNA. Breaking one strand generates a nick and breaking both strands generates a double-strand break.
An endonuclease cuts in the middle of RNA or DNA, while an exonuclease cuts from the end of the strand. Genome engineering tools like Cas9 are endonucleases.
-
Nucleic acidA term for DNA and RNA. Refers to nucleotides, the basic chemical units that are strung together to make DNA or RNA. One of the four macromolecules that make up all living things (protein, lipids, carbohydrates, and nucleic acids).
-
NucleotideOne of the basic chemical units strung together to make DNA or RNA. Consists of a base, a sugar, and a phosphate group. The phosphates can link with sugars to form a string called the DNA/RNA backbone, while the bases can bind to their complementary partners to form base pairs.
-
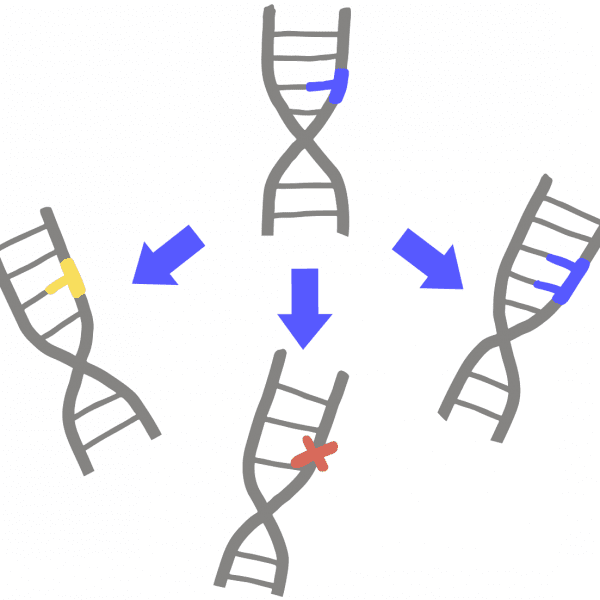
Off-target effectWhen a genome engineering enzyme cuts DNA at an unintended, "off-target," site that is similar to the intended target.
-
On-target effect
When a genome-editing enzyme cuts DNA at the intended ”on-target” site, but makes a different change to the DNA than desired.
-
Protospacer adjacent motif (PAM)A short sequence that must be present next to a DNA target sequence for Cas9 to bind and cut. Prevents cleavage of host CRISPR array, where PAM is not present.
-
PathogenA microbe that causes illness. Most micro-organisms are not pathogenic to humans, but some strains or species are harmful.
-
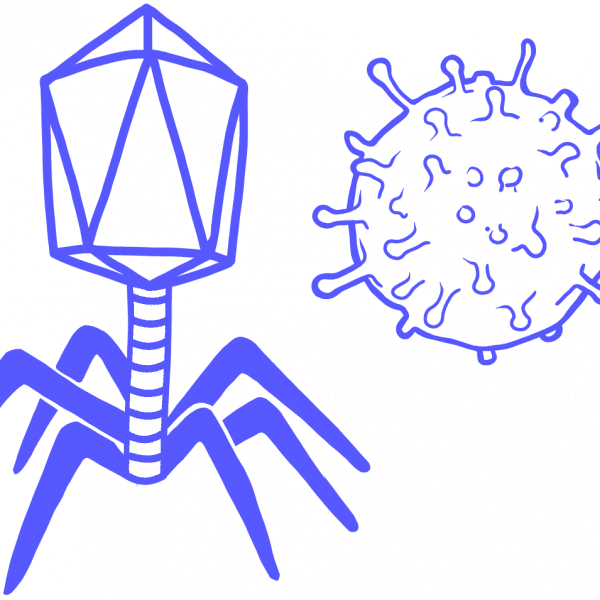
PhageA type of virus that infects bacteria or archaea, formally called bacteriophage.
-
Plasmid
A circular piece of DNA found naturally in bacteria, often used to carry new genetic material into cells.
-
Polygenic
Describes a trait that is shaped by the combined effect of multiple genes. Compare to monogenic
-
Prime editing
A type of genome editing that uses CRISPR components to directly change any single DNA letter to any other letter or make small insertions without making breaks in the DNA. Prime editors are made by fusing (attaching) DNA-modifying enzymes to DNA-targeting Cas proteins like Cas9.
-
ProkaryoteA category of living organisms that encompasses all bacteria and archaea. Prokaryotes are microscopic, single-celled organisms that do not have a nucleus or other membrane-bound organelles. Compare to eukaryotes.
-
ProteinA string of amino acids folded into a three-dimensional structure. Proteins are each specialized to perform a specific role to help cells grow, divide, and function. One of the four macromolecules that make up all living things (protein, lipids, carbohydrates, and nucleic acids).
-
Ribonucleoprotein complex (RNP)An assembly of molecules containing both protein and RNA. Often used to describe Cas9 protein bound to guide RNA (gRNA), which together form an active enzyme. For genome editing in cells, Cas9 can be delivered as a pre-assembled RNP, or as DNA or RNA encoding the genetic instructions for the protein and RNA components.
-
RNAAbbreviation of ribonucleic acid. Transcribed from a DNA template and typically used to direct the synthesis of proteins. CRISPR-associated proteins use RNAs as guides to find matching target sequences in DNA.
-
SARS-CoV-2
Short for severe acute respiratory syndrome coronavirus 2. SARS-CoV-2 is the virus that causes COVID-19.
-
Single-guide RNA (sgRNA)A version of the naturally occurring two-piece guide RNA complex engineered into a single, continuous sequence. The simplified single-guide RNA is used to direct the Cas9 protein to bind and cleave a particular DNA sequence for genome editing.
-
Somatic cellsAll the cells in a multicellular organism except for germ cells (eggs or sperm). Mutations or changes to the DNA in the soma will not be inherited by subsequent generations.
-
Stakeholder
Someone who can take an action on a goal or project, or who will be affected by those actions. Stakeholders can be individuals or groups of people.
-
Stem cellsCells with the potential to turn into a specialized type of cell or to divide to make more stem cells. Most cells in your body are differentiated – that is, their fate has already been decided and they cannot morph into a different kind of cell. For example, a cell in your brain cannot transform into a skin cell. Embryonic stem cells are found in developing embryos, while adult stem cells are found in tissues including bone marrow, blood, and fat. Adult stem cells replenish the body as it becomes damaged over time.
-
StrandA string of connected nucleotides; can be DNA or RNA. Two strands of DNA can zip together when complementary, bases match up to form base pairs. DNA typically exists in this double-stranded form, which takes the shape of a twisted ladder or double helix. RNA is typically composed of just a single strand, though it can fold up into complex shapes.
-
T cells
A type of white blood cell that helps protect the body from infection and cancer. Can be genetically engineered to better fight cancer.
-
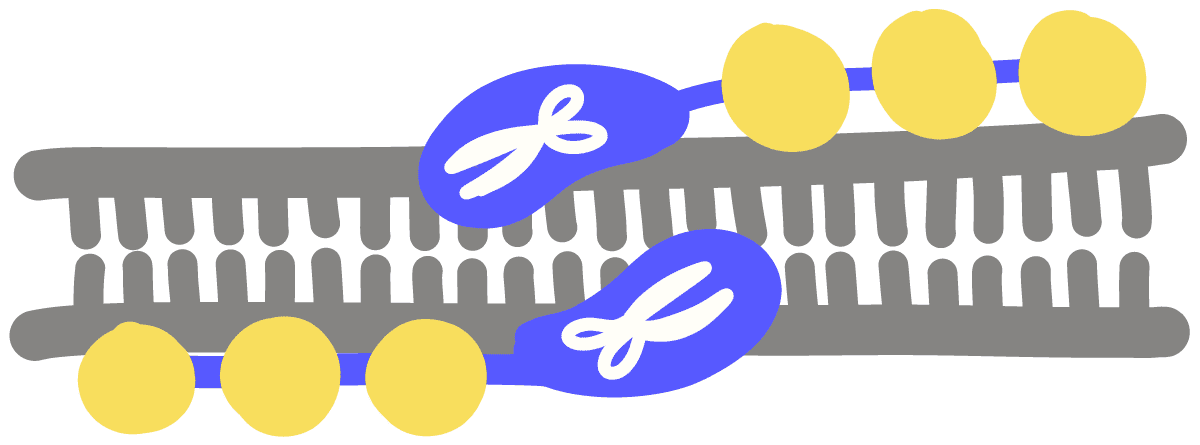
Transcription activator-like effector nuclease (TALEN)A genetic engineering tool wherein one portion of the protein recognizes a specific DNA sequence and another part cuts DNA. Made by attaching a series of smaller DNA-binding domains together to recognize a longer DNA sequence. This DNA-binding domain is fused to a nuclease that will cut nearby DNA. Like CRISPR-Cas9 and ZFNs, it can be used to alter DNA sequences.
-
Trans-activating CRISPR RNA (tracrRNA)Abbreviation for trans-activating CRISPR RNA, pronounced “tracer RNA.” In the CRISPR-Cas9 system, the tracrRNA base pairs with the crRNA to form a functional guide RNA (gRNA). Cas9 uses the tracrRNA portion of the guide as a handle, while the crRNA spacer sequence directs the complex to a matching viral sequence.
-
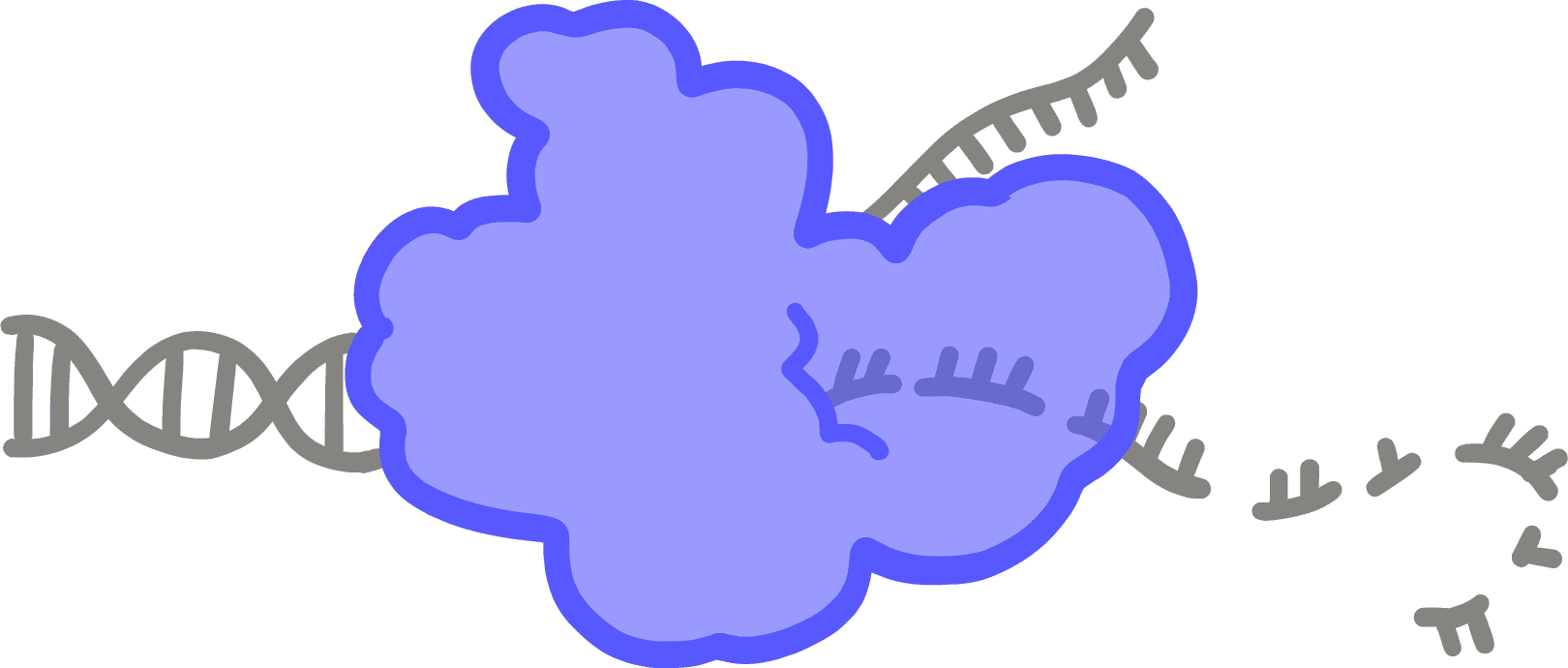
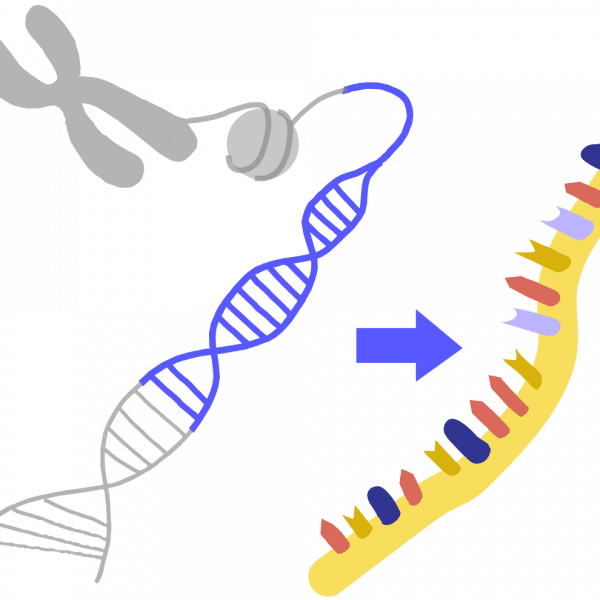
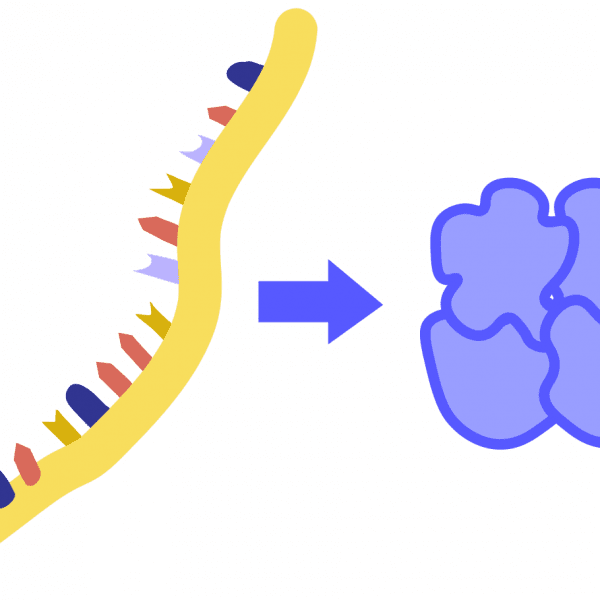
TranscriptionThe process by which DNA information is copied into a strand of RNA; performed by an enzyme called RNA polymerase.
-
Transgene
A gene from one organism that is put into a different organism through genome engineering.
-
TranslationThe process by which proteins are made based on instructions encoded in an RNA molecule. Performed by a molecular machine called the ribosome, which links together a series of amino acid building blocks. The resulting polypeptide chain folds up into a particular 3D shape, known as a protein.
-
Vaccine
A treatment used to stimulate the body’s immune system to provide protection from a specific disease.
-
Viral vector
A virus engineered to deliver genes or genome-editing molecules without causing infection or disease. Adeno-associated virus (AAV) and lentivirus (LV) are common examples.
-
VirusAn infectious entity that can only persist by hijacking a host organism to replicate itself. Has its own genome, but is technically not considered a living organism. Viruses infect all organisms, from humans to plants to microbes. Multicellular organisms have sophisticated immune systems that combat viruses, while CRISPR systems evolved to stop viral infection in bacteria and archaea.
-
Zinc-finger nuclease (ZFN)A genetic engineering tool wherein one portion of the protein recognizes a specific DNA sequence and another part cuts DNA. Made by attaching a series of smaller DNA-binding domains together to recognize a longer DNA sequence. This DNA-binding domain is fused to a nuclease that will cut nearby DNA. Like CRISPR-Cas9 and TALENs, it can be used to alter DNA sequences.
Use and Attribution:
We encourage you to use and adapt these images for non-commercial, educational purposes. Tweet us @igisci to show us how you’re using them! Download editable image files by clicking here. ALL modifications and uses must follow the CC BY-NC-SA 4.0 license:
This IGI Glossary Icon Collection by Christine Liu of Two Photon Art for the Innovative Genomics Institute is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.