Missing Link Found in the Evolutionary History of Carbon-Fixing Protein Rubisco
The enzyme rubisco is found in all plants and other photosynthesizing organisms. It plays a key role in fixing carbon from the air and has helped shape life on Earth. Now researchers at UC Davis, the Innovative Genomics Institute at UC Berkeley, and the Lawrence Berkeley National Lab have identified a version of rubisco from environmental microorganisms dating back billions of years.
Missing Link Found in the Evolutionary History of Carbon-Fixing Protein Rubisco
UC Davis News | Greg Watry | August 31, 2020
A team led by researchers at the University of California, Davis has discovered a missing link in the evolution of photosynthesis and carbon fixation. Dating back more than 2.4 billion years, a newly discovered form of the plant enzyme rubisco could give new insight into plant evolution and breeding.
Rubisco is the most abundant enzyme on the planet. Present in plants, cyanobacteria (also known as blue-green algae), and other photosynthetic organisms, it’s central to the process of carbon fixation and is one of Earth’s oldest carbon-fixing enzymes.
“It’s the primary driver for producing food, so it can take CO2 from the atmosphere and fix that into sugar for plants and other photosynthetic organisms to use. It’s the primary driving enzyme for feeding carbon into life that way,” said Doug Banda, a postdoctoral scholar in the lab of Patrick Shih, Assistant Professor of Plant Biology in the UC Davis College of Biological Sciences.
Form I rubisco evolved over 2.4 billion years ago before the Great Oxygenation Event, when cyanobacteria transformed the Earth’s atmosphere by producing oxygen through photosynthesis. Rubisco’s ties to this ancient event make it important to scientists studying the evolution of life.
In a study appearing Aug. 31 in Nature Plants, Banda and researchers from UC Davis, the Innovative Genomics Institute (IGI), and the Lawrence Berkeley National Laboratory report the discovery of a previously unknown relative of form I rubisco, one that they suspect diverged from form I rubisco prior to the evolution of cyanobacteria.
The new version, called form I-prime rubisco, was found through genome sequencing of environmental samples and synthesized in the lab. Form I-prime rubisco gives researchers new insights into the structural evolution of form I rubisco, potentially providing clues as to how this enzyme changed the planet.
An Invisible World
Form I rubisco is responsible for the vast majority of carbon fixation on Earth. But other forms of rubisco exist in bacteria and in the group of microorganisms called Archaea. These rubisco variants come in different shapes and sizes, and even lack small subunits. Yet they still function.
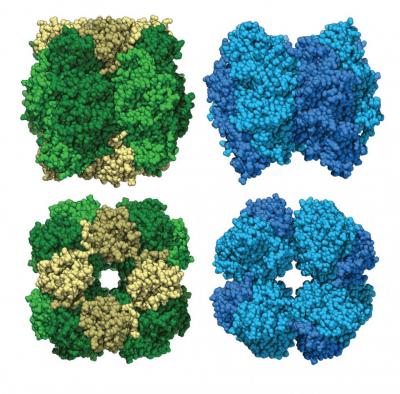
“Something intrinsic to understanding how form I rubisco evolved is knowing how the small subunit evolved,” said Shih. “It’s the only form of rubisco, that we know of, that makes this kind of octameric assembly of large subunits.”
Study co-author Jill Banfield, IGI Investigator and UC Berkeley Professor of Earth and Planetary Science, uncovered the new rubisco variant after performing metagenomic analyses on groundwater samples. Metagenomic analyses allow researchers to examine genes and genetic sequences from the environment without culturing microorganisms.
“We know almost nothing about what sort of microbial life exists in the world around us, and so the vast majority of diversity has been invisible,” said Banfield. “The sequences that we handed to Patrick’s lab actually come from organisms that were not represented in any databases.”
Banda and Shih successfully expressed form I-prime rubisco in the lab using E. coli and studied its molecular structure.
Form I rubisco is built from eight core large molecular subunits with eight small subunits perched on top and bottom. Each piece of the structure is important to photosynthesis and carbon fixation. Like form I rubisco, form I-prime rubisco is built from eight large subunits. However, it does not possess the small subunits previously thought essential.
“The discovery of an octameric rubisco that forms without small subunits allows us to ask evolutionary questions about what life would’ve looked like without the functionality imparted by small subunits,” said Banda. “Specifically, we found that form I-prime enzymes had to evolve fortified interactions in the absence of small subunits, which enabled structural stability in a time when Earth’s atmosphere was rapidly changing.”
According to the researchers, form I-prime rubisco represents a missing link in evolutionary history. Since form I rubisco converts inorganic carbon into plant biomass, further research on its structure and functionality could lead to innovations in agriculture production.
“Although there is significant interest in engineering a ‘better’ rubisco, there has been little success over decades of research,” said Shih. “Thus, understanding how the enzyme has evolved over billions of years may provide key insight into future engineering efforts, which could ultimately improve photosynthetic productivity in crops.”
Additional authors on the study are: Albert Liu at UC Davis and LBNL; Jose Pereira and Paul Adams, Joint BioEnergy Institute, LBNL; Christine He, UC Berkeley; Michal Hammel, LBNL; and Douglas Orr, Martin Parry and Elizabete Carmo-Silva, Lancaster University, U.K. The work was partly supported by the U.S. Department of Energy, Branco Weiss Fellowship, the Biotechnology and Biological Sciences Research Council (U.K.), NIH, the Chan Zuckerberg Biohub, and the Innovative Genomics Institute.
Media Contact
Andy Murdock: andymurdock@berkeley.edu