CRISPR-based gene drive architecture for control of agricultural pests
Project Overview
We are using mathematical models to determine ideal strategies for CRISPR-Cas9-based gene drive systems to control insect crop pests responsible for major losses in global food production.
The advent of CRISPR-Cas9 technology has greatly increased the ease of genome editing. One potential application of this is the engineering of gene drive systems capable of spreading desirable genes into populations and/or suppressing populations. Initial applications of this technology have been discussed in the context of controlling mosquitoes and the diseases they transmit; however, the technology applies broadly to species with short generation times that reproduce sexually, including agricultural insect pests. Between 10–16% of potential global food production is lost to insect pests every year. Hence, agricultural applications of gene drive technology could provide significant societal benefits by enhancing food security and reducing reliance on environmentally destructive insecticides.
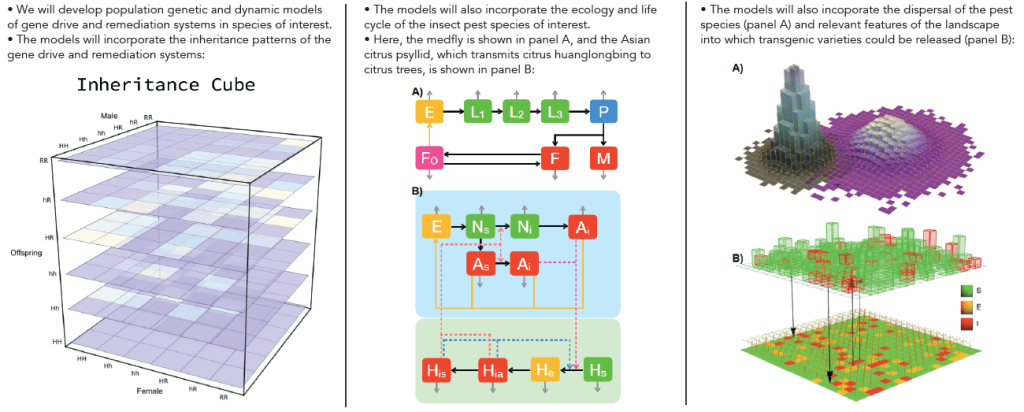
Our lab has substantial experience developing mathematical models of gene drive systems for the purpose of controlling mosquito populations and the diseases they transmit. The goal of this project is to leverage this expertise in order to apply it to pest species of agricultural significance. We have identified a variety of crop pests of interest to which this technology may apply: i) Drosophila suzukii, a worldwide pest that infests fruit during the ripening stage; ii) the medfly, a worldwide pest native to the Mediterranean that infests several species of fruits; iii) the pink bollworm, an invasive pest in most of the world’s cotton-growing regions; and iv) the Asian citrus psyllid, a vector of citrus huanglongbing virus present in Asia and the southern US. For each of these species, we will use mathematical models to determine optimal CRISPR-Cas9-based gene drive architectures that could be successful in controlling their agricultural impact while ensuring biosafety through the ability to remediate them from the environment in the event of negative consequences or a change in public opinion.
Principal Investigators
- John Marshall
Publications
Manipulating the Destiny of Wild Populations Using CRISPR
Raban R, Marshall JM, Hay BA, and Akbari OS.
Genetic conversion of a split-drive into a full-drive element
Terradas G, Bennett J, Li Z, Marshall JM, and Bier E. Nature Communications
Exploring the value of a global gene drive project registry
Taitingfong RI, Triplett C, Vásquez VN … Marshall JM, Montague M, Morrison AC, Opesen CC, Phelan R, Piaggio A, Quemada H, Rudenko L, Sawadogo N, Smith R, Tuten H, Ullah A, Vorsino A, Windbichler N, Akbari OS, Long K, Lavery JV, Evans SW, Tountas K, Bloss CS. Nature Biotechnology
Precision guided sterile males suppress populations of an invasive crop pest
Kandul NP, Liu J, Buchman A, Shriner IC, Corder RM, Warsinger-Pepe N, Yang T, Yadav AK, Scott MJ, Marshall JM, and Akbari OS. GEN Biotechnology
Exploiting a Y chromosome-linked Cas9 for sex selection and gene drive
Gamez S, Chaverra-Rodriguez D, Buchman A, Kandul NP, Mendez-Sanchez SC, Bennett JB, Sánchez C. HM, Yang T, Antoshechkin I, Duque JE, Papathanos PA, Marshall JM, and Akbari OS. Nature Communications
Winning the tug-of-war between effector gene design and pathogen evolution in vector population replacement strategies.
Marshall JM, Raban RR, Kandul NP, Edula JR, León TM, and Akbari OS. Frontiers in Genetics
MGDriveE: A modular simulation framework for the spread of gene drives through spatially-explicit mosquito populations.
Sánchez HM, Wu SL, Bennett JB, and Marshall JM. Methods in Ecology and Evolution
Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management.
KaramiNejadRanjbar M, Eckermann KN, Ahmed HMM, Sánchez C HM, Dippel S, Marshall JM, and Wimmer EA. PNAS
Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii.
Buchman A, Marshall JM, Ostrovski D, Yang T, and Akbari OS. PNAS. Preprint (free to read).
