IGI Researchers Increase CRISPR-Cas9 Efficiency
CRISPR-Cas9 breaks genes better if you disrupt DNA repair
Berkeley News | Robert Sanders | August 17, 2016
CRISPR-Cas9 is the go-to technique for knocking out genes in human cell lines to discover what the genes do, but the efficiency with which it disables genes can vary immensely.
UC Berkeley researchers have now found a way to boost the efficiency with which CRISPR-Cas9 cuts and disables genes up to fivefold, in most types of human cells, making it easler to create and study knockout cell lines and, potentially, disable a mutant gene as a form of human therapy.
Scientists are constantly discovering new genes or the proteins they code for, but it’s much harder to figure out their role in the body or in disease. Key to discovering this role is disabling the gene to see what happens when it’s removed.
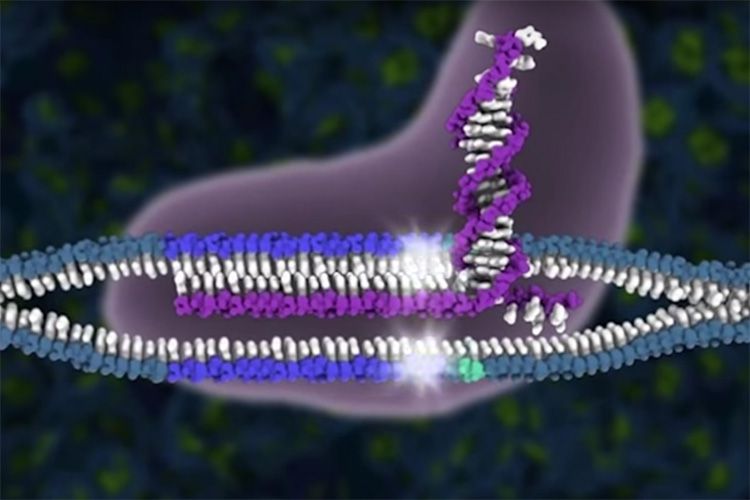
CRISPR-Cas9 cuts DNA: The CRISPR-Cas9 complex (purple) cuts a targeted piece of DNA repeatedly until the cell’s repair enzymes make a mistake in patching the cut, effectively disabling the gene. UC Berkeley scientists achieved much better cutting success by interfering with the DNA repair process, boosting the likelihood of a botched patch. This graphic shows the guide RNAs (purple) that pair with complementary DNA (blue) and align the Cas9 protein to precisely cut the two strands of DNA (white stars).
While CRISPR-Cas9 can accelerate the process of making knockout cell lines, researchers must sometimes make and screen many variations of the genetic scissors to find one that works well. The UC Berkeley researchers found that this process can be made many times more efficient with a simple tweak.
The key is introducing into the cell, along with the CRISPR-Cas9 protein, short pieces of DNA that do not match any DNA sequences in the human genome. The short pieces of DNA, called oligonucleotides, seem to interfere with the DNA repair mechanisms in the cell to boost the editing performance of even mediocre CRISPR-Cas9s between 2½ and 5 times.
“It turns out that if you do something really simple — just feed cells inexpensive synthetic oligonucleotides that have no homology anywhere in the human genome — the rates of editing go up as much as five times,“ said lead researcher Jacob Corn, the scientific director of UC Bekeley’s Innovative Genomics Initiative and an assistant adjunct professor of molecular and cell biology. The technique boosts the efficiency of all CRISPR-Cas9s, even those that initially failed to work at all.
With higher efficiency, researchers will have better success at creating the knockouts they want, and then using those knockout cell lines to explore the function of a gene or a group of genes. Because most long-lived cell lines are derived from cancer cells — including the very popular HeLa cell line — these cell lines typically have more than the normal two copies of each gene. This can make it difficult to knock out all copies at once, and higher efficiency greatly increases the chance of success.
High efficiency also is essential when knocking out genes to correct hereditary mutations in humans. Physicians have speculated about knocking out genes that make people susceptible to infectious diseases, such as AIDS, or prone to autoimmune, inflammatory or neurodegenerative disorders. It remains to be seen whether the approach described by Corn and colleagues could be used in a therapeutic context, but it is very effective for research purposes.
The results will be reported Aug. 17 in the online journal Nature Communications.
DNA repair key to CRISPR-Cas9 success
The CRISPR-Cas9 molecule consists of a protein scissors, the Cas9 protein, and an address telling Cas9 where to bind the DNA and cut. The technique relies upon the fact that when you cut DNA, the cell’s repair mechanisms don’t always correctly rebuild the DNA strand, but make an error in the sequence that disables the gene and knocks out its activity.
Tha address, called a guide RNA, is a string of 20 ribonucleic acid molecules that are complementary to the DNA sequence of the target gene. This guide RNA binds to the DNA like a strip of Velcro, setting up Cas9 to cut the double-stranded DNA.
Why some guide RNAs work well, setting up Cas9 to cut and disable a gene nearly 100 percent of the time, while others bind but seldom or never knock out the gene, has been a puzzle since the technique was invented by Jennifer Doudna of UC Berkeley and Emmanuelle Charpentier of Umea University in 2012. The cutting efficiency varies with the type of cell and the particular cell line, Corn said.
Corn suspected that Cas9’s occasionally poor cutting efficiency might be related to how DNA is repaired, since DNA repair mechanisms — the basic housekeeping enzymes that fix any breaks or deletions in the DNA that might lead to a deadly mutation — differ from cell to cell. He reasoned that random strands of DNA — none of them similar to any actual human DNA (that is, non-homologous) — might confuse the repair process and improve the knockout success rate.
“It gives the cell a little kick to prevent normal repair from happening,“ he said.
He portrays CRISPR-Cas9 gene editing as a competition between cutting and DNA repair: once Cas9 cuts, the cell exactly replaces the cut DNA, which Cas9 cuts again, in an endless cycle of cut and repair until the repair enzymes make a mistake and the gene ends up disfunctional. Perhaps, he said, the oligonucleotides decrease the fidelity of the repair process, or make the cell switch to a more error-prone repair that allows Cas9 to more readily break the gene.
The next frontier, he said, is trying to take advantage of the peculiarities of DNA repair to improve sequence insertion, in order to replace a defective gene with a normal gene and possibly cure a genetic disease.
The research, co-authored by postdoctoral fellows Chris Richardson and Nicolas Bray and former research associate Jordan Ray, was funded by the Li Ka Shing Foundation.
—
Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes
Nature Communications | CD Richardson, GJ Ray, NL Bray & JE Corn | August 17, 2016
You may also be interested in

Wolf Prize Laureate Brian Staskawicz on 40 Years of Plant Immunity Research

Announcing the Rising Stars Program: A New Collaboration Between the IGI and Historically Black Colleges and Universities

Breakthrough Method Enables Rapid Discovery of New Useful Proteins
