CRISPR Diversifies: Cut, Paste, On, Off, and Now– Evolve!
Life is astoundingly diverse. By taking antibiotics to stop infections or using yeast to brew beer, we are co-opting useful products and processes that evolved naturally. But what happens when the trait we want can’t be found in nature?
Scientists at the Innovative Genomics Institute have concocted a transformative new way to harness the power of evolution. Today in Nature, researchers led by PhD student Shakked Halperin working in the laboratories of David Schaffer and John Dueber at UC Berkeley describe yet another creative application for CRISPR: a platform to spur evolution of specific genes inside cells. Their inventive new system, “EvolvR,” lets scientists shake up the DNA letters in their gene of choice until they find the variation that’s just right. The technology opens up countless possibilities, like engineering yeast that efficiently turn waste into biofuels, or developing new human therapeutics.
Not just monkeying around
Imagine a monkey sitting at a keyboard. If given an infinite amount of time to press keys at random, the monkey would almost surely type the complete work of William Shakespeare.
At least, that’s according to the “infinite monkey theorem.” Natural DNA variation is akin to this process – as time goes by, random changes pop up across the genomes of different individual organisms.
In theory, over infinite time, every possible variation of the genetic letters will have existed. On a practical human timeline, however, only a very small fraction of possible variations will ever appear.
Now imagine that we can tell the monkey to rewrite just a specific page in Shakespeare’s Macbeth. Limited to just this narrow window, the monkey will type out every possible variation of the page’s text much, much faster. This is what EvolvR lets scientists do. They only want new versions of a single gene, so rewriting the entire genome is impractical and likely toxic to living cells. By limiting the scrambling to just one gene at a time, it becomes possible to sample an enormous number of variations.
Flicking on the “evolve” switch
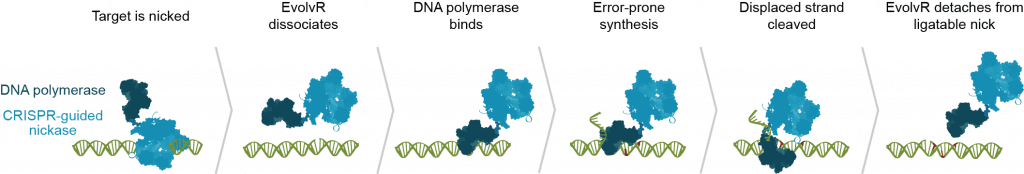
EvolvR lets scientists push a gene through the entire process of evolution in just one day in the lab. The system is built on the programmable DNA cutting protein Cas9, making EvolvR the latest ingenious device in the CRISPR toolbox. The IGI team leashed Cas9 to an enzyme called DNA polymerase. Cas9 is programmed to find a specific target sequence in an organism’s DNA. EvolvR uses a special “nicking” version of Cas9 that only cuts one of the two DNA strands. Cas9 makes a nick, signaling for DNA polymerase to peel back the strand and replace it with new DNA. The polymerase makes errors, writing in a different DNA sequence than the original, like the proverbial monkey at the keyboard.
Since diversity is the goal, the polymerase’s “typos” are a good thing. Scientists can use EvolvR to deliberately make random mutations, creating millions of different sequence combinations and likely finding at least one that has the effect they want.
Directed evolution technology evolves
This method is a fundamentally new way to diversify biological systems, opening doors that remained locked with earlier strategies. Other methods relied on forcing a vast “library” of randomized DNA pieces into cells. This is time-consuming, expensive, and not all cells will easily take up external DNA. EvolvR gets around these and several other drawbacks of previous approaches. “It doesn’t require a double-strand break like many other technologies do,” notes Dueber. “Double-strand breaks are toxic to many cells. It also doesn’t require sophisticated repair pathways, which many interesting organisms don’t have.”
The tool should therefore work in any species, and testing this idea is one of the team’s immediate next steps. Though its proving ground was in bacteria, the versatile platform will be even more powerful when used in eukaryotes, like human or plant cells. “EvolvR has huge potential as a species-independent tool for directed evolution,” says Schaffer. “It can already make single mutations or combinations of changes across regions of DNA ranging from about a dozen to a few hundred base pairs long.” Dueber adds, “We’re interested in making a modest toolkit of EvolvRs. We envision systems that have higher mutation rates or that affect much larger windows. There are a lot of ideas to try.”
Halperin points out another key strength of the system. Earlier methods for evolving traits in the lab include just a few, labor-intensive rounds of diversification and selection. “In previous directed evolution experiments, we stepped away from what nature does. Because our tool can continuously provide diversity at a target gene, we can continuously enrich for better and better traits, closer to the natural evolutionary process.” An EvolvR experiment can go on as long as the researcher wants, shuffling the gene’s sequence over and over again and creating more opportunities for success.
Off to the races
There are almost too many possibilities to envision up front, and the team behind EvolvR hopes that other scientists will take their creation and run with it.
“We’re excited about other people joining us to use this tool and improve it,” says Halperin. Schaffer is eager to implement the tool to “accelerate the development of biomolecules for human therapeutic applications, from new drugs to new drug delivery technologies.”
The researchers are especially excited about combining EvolvR with another CRISPR toolbox favorite, high-throughput CRISPR screening. This potent pairing of technologies would let them diversify thousands of different genes in a single experiment, potentially creating brand new functions instead of just turning genes on and off.
A single drop of bacteria diversified with EvolvR contains immense diversity. What groundbreaking potential might be hiding in that single drop? As Kahlil Gibran once wrote, “In one drop of water are found all the secrets of all the oceans.” While Gibran’s poetic vision is not quite realized, we are quickly approaching it.
—
This study was published in the online edition of the journal Nature as “CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window.” In addition to Dueber, Halperin, and Schaffer, co-authors of the work are Connor Tou, Eric Wong, and Cyrus Modavi. This research was funded by the Innovative Genomics Institute.
The Innovative Genomics Institute (IGI) is a non-profit, academic partnership between UC Berkeley and UC San Francisco that supports collaborative research projects across the Bay Area. The IGI’s mission is to develop and deploy genome engineering to cure disease, ensure food security, and sustain the environment for current and future generations. As pioneers in genome editing, functional genomics, and other cutting-edge technologies, IGI scientists continuously push the boundaries of science.
Media Contacts
John Dueber: jdueber@berkeley.edu
Shakked Halperin: shakked@berkeley.edu
David Schaffer: schaffer@berkeley.edu
You may also be interested in
T Cell Engineering Breakthrough Sidesteps Need for Viruses in Genome Editing

Mike Lenardo Seminar: Precision Medicine Treatments from Identification of Defects in Immune Disorders
IGI Seminar Series: Progress and Challenges in Delivering Cassava with RNAi-mediated Resistance to Cassava Brown Streak Disease to Smallholder Farmers in Africa – It’s Not Just About the Technology



 By
Megan Hochstrasser
By
Megan Hochstrasser