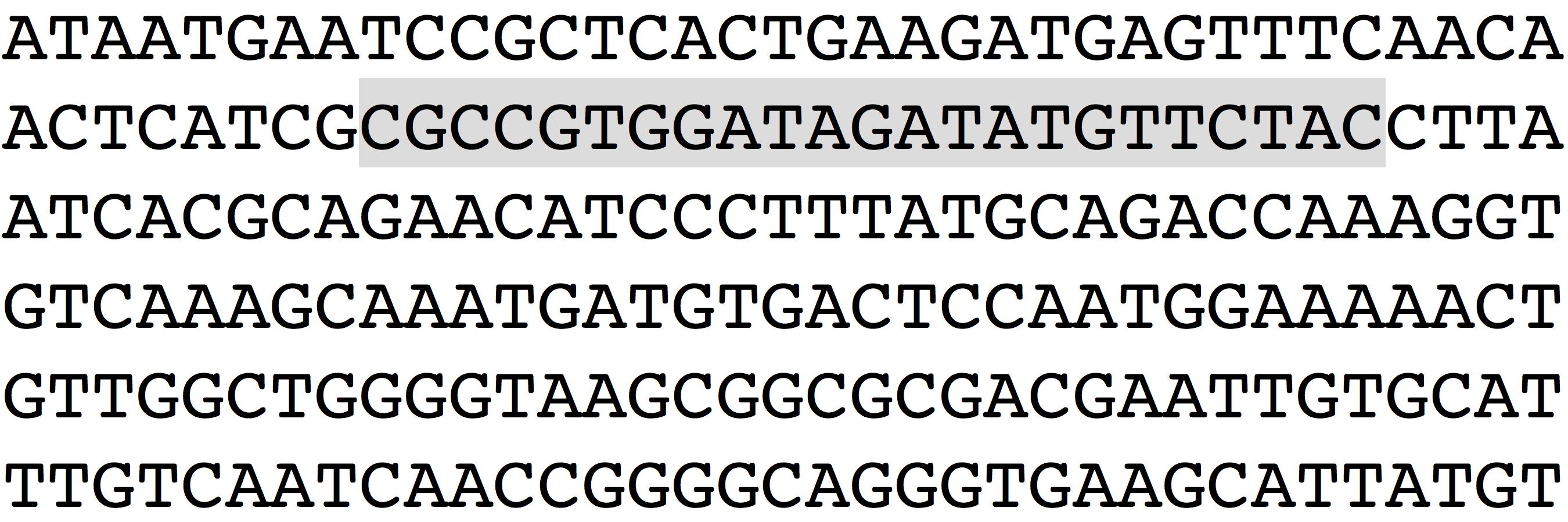
How to Make a Guide RNA for a Cas9 Knockout
Guide RNA lore is split across multiple papers, people, and places, and I’m frequently asked about the “best” way to make a guide RNA for Cas9. The following is the state of the art as I understand it, as of today (8/11/14), split into several steps. The steps below assume you want to use Streptococcus pyogenes Cas9 to cut a gene to introduce an insertion/deletion (“indel”) to make a knockout (the simplest use case) using a double-strand cut (wild type Cas9). The process may differ if you want to (for example) use CRISPRi to inhibit transcription. I’ve used * to mark steps that would be at least somewhat altered for other applications or if you’re using less common parts (e.g. Cas9 from another species, different guide RNA promoter, etc). Before you start
- Decide what kind of targeting you want to do. Here we’re considering double-stranded cutting to make a knockout via introduction of an indel.
- Decide which Cas9 you’ll use. Here, we’ll assume you’re using Streptococcus pyogenes (aka “Spy“). This choice would affect the protospacer adjacent motif (“PAM”) you’ll look for.*
- Get the genomic sequence you want to target from NCBI Gene or elsewhere (e.g. if you’re targeting an intergenic region).*
- For knockouts, you generally want to introduce an indel as close to the 5′ end of the coding region as possible. This will have the highest likelihood of creating a protein-destroying frameshift.*
Finding Guides
- Use one of many servers to find guide RNAs in the region you’d like to cut. For example, CRISPR-MIT, E-CRISP, or CHOPCHOP. Which tool you choose is mostly personal preference, and each has their own model for scoring guide RNAs.
- Each target site will either be ~23 bases ending in “
GG” (guide binds Crick strand) or ~23 bases starting in “CC” (guide binds Watson strand). The protospacer adjacent motif (“PAM”) refers to those last or first three bases and is present in the DNA you’re targeting but should not be used in the guide RNA. Hence, the guide RNA itself will be ~20 bases and lack a PAM. SpyCas9 can also use “AG” (Watson “CT“) as a PAM, but not as efficiently. - The exact length of the guide doesn’t seem to matter very much; anything from 17-27 bases (remember, guides don’t include the PAM) seems basically OK (with some qualifiers).
Choosing a guide Now you have a (possibly very long) list of potential guides. Each one has an associated score. How do you choose which one to use? Here’s a semi-ordered list of factors to consider, from most to least important. Consider these qualitative, rather than a quantitative score.
- For a knockout, it doesn’t matter which strand the guide RNA binds, but CRISPRi guides should be complementary to the non-coding strand.*
- Guides should be perfectly complementary to the region you want to target in the 8-12 bases closest to the PAM.
- Never choose a guide that has any significant off-target sites (perfect match for the 8-12 bases closest to the PAM) in a coding of the genome.
- Never use a guide with >=3 U’s in a row, since these sequences act as Pol III terminators. This is obviously not applicable if you are using a Pol II vector instead of the common U6 promoter vectors.*
- Prefer guides with a PAM of
NGGinstead ofNAG.* - Avoid sequences with significant secondary structure (The Vienna web server is a great place to check this). You should also avoid guides that disrupt the secondary structure of the 3′ constant region (the most common constant region sequence is
"GUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUUUU"). - GC content should be between ~ 30-80%, the higher the better (but not too high!).
- Avoid additional
G‘s after the PAM. For example, a genomic sequence of “|AGG|CCAT” is probably OK (where “AGG” is the PAM). But “|AGG|GCAT” is probably not a good idea, and “|AGG|GGGG” is a definite no-no.* - Some groups have shown that
U‘s are disfavored in the -1, -2, and -4 position (counting back from the first base of the constant region). Other groups haven’t seen this. Your mileage may vary. - Prefer guides in DNAse hypersensitive regions (as annotated by ENCODE on the UCSC genome browser). This isn’t a necessity, but probably won’t hurt.
- It’s recently been shown that microhomology at the site of cutting can substantially increase the chance of getting an out-of-frame indel. This doesn’t affect cutting itself, but could help you get the knockout.
Construction of the final guide
- Take the guide sequence you chose above and append the constant sequence “
GUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUUUU” to the 3′ end. - If your guide does not begin with a 5′
G, just add one. This increases efficiency of transcription from the U6 promoter and does not need to be homologous to the region you’re targeting.* - Add cloning sites appropriate for the expression vector you’ve chosen. For example, if using the Zhang’s lab pX330 vector, append “
CACC” to the 5′ end of the Watson strand and “AAAC” to the 5′ end of the Crick strand.* - Order oligos, anneal, and ligate.
The above might seem like a lot, but it’s really not all that bad. You’ll quickly get a feel for what makes good vs bad guides. Since it’s so easy to test multiple guides, I always recommend making at least two guides per knockout you’d like to make. That way if one is a dud, you aren’t caught flat-footed. Obviously, there are many *s in the list above, denoting steps that might be a bit different if your application or parts differ from SpyCas9 making a double-strand break for the purposes of a knockout. The toolbox is always expanding, so options abound! But hopefully the above provides a general idea of how to get started. Do you have another neat trick to share? Did I miss something important? Want to expound on the best way to make a CRISPRi guide (a whole other ball of wax)? Feel free to leave a comment!
You may also be interested in
IGI Seminar Series: Progress and Challenges in Delivering Cassava with RNAi-mediated Resistance to Cassava Brown Streak Disease to Smallholder Farmers in Africa – It’s Not Just About the Technology

IGI Seminar Series: Empowering Teachers, Transforming Classrooms: Advancing K-12 STEM Education

IGI Seminar Series: Writing DNA with RNA: Genome Engineering by Target-Primed Reverse Transcription

 By
Jacob Corn
By
Jacob Corn