Genomics
Institute
Structural studies of the COVID-19 (SARS-CoV-2 virus) replication complex
We are doing a structural investigation into the replication of SARS-CoV-2 in order to find new targets for COVID-19 treatments.
SHARE:
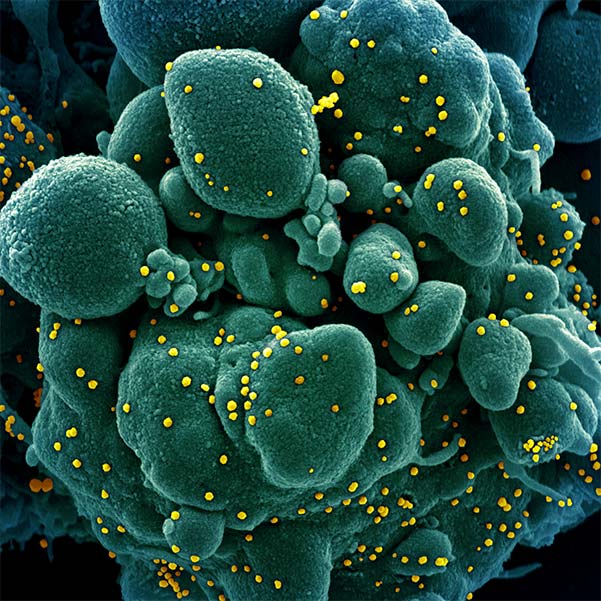
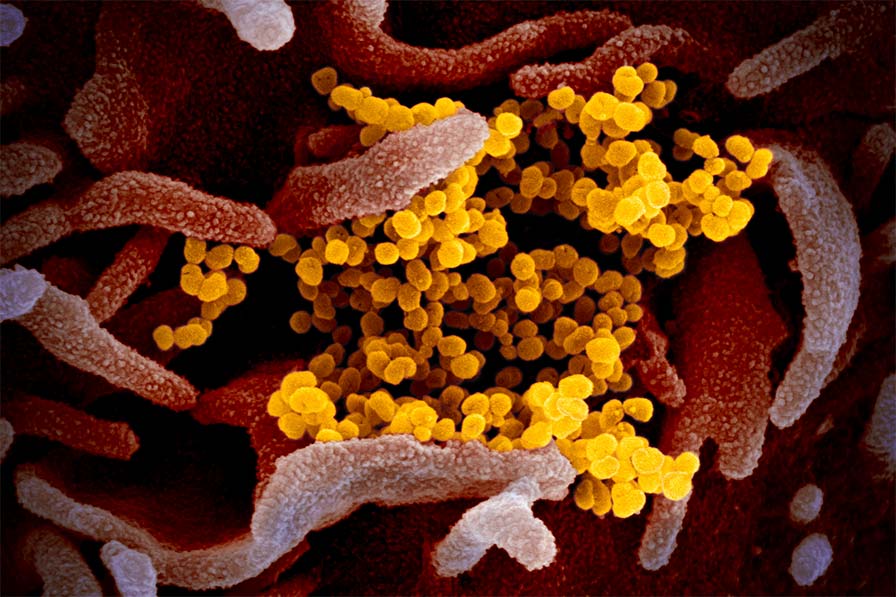
SARS-CoV-2 uses an RNA-dependent RNA polymerase complex (RdRp) to both replicate and transcribe the viral genome. Because this complex is crucial for viral propagation, it is a prime candidate as a drug target against COVID-19. In addition to the core RdRp that includes the RNA polymerase NSP12 and the fidelity factors NSP7/8, the holo-complex includes several other viral proteins that play critical roles in synthesis fidelity and RNA processing: (i) the virus maintains its large genomes by encoding a proofreading complex of NSP10/14 that removes mis-incorporated nucleotides; (ii) the mRNA synthesized by the RdRp requires a mGpppA cap to be translated by the host ribosome, so NSP10/14 first caps the RNA with a GpppA cap and then NSP10/16 methylates it; (iii) NSP13 plays a role in separating the newly synthesized product strand from the template strand, while NSP9 stabilizes the two single-stranded RNAs.
Our goal is to reconstitute and determine the structure of the SARS-CoV-2 RdRp holo-complex during the stages of initiation of RNA synthesis, capping, and proofreading using cryo-electron microscopy in order to better understand how SARS-CoV-2 replicates and transcribes its genome. This mechanistic level of understanding will open new doors for the development of drugs to fight COVID-19 that exploit new sites for drug binding and complex inactivation.
Share this project: