Genomics
Institute
Discovering and targeting the RNA structural code underlying COVID-19
We are developing computational and experimental tools to identify and target SARS-CoV-2 RNA structural elements that will lead to new COVID-19 therapies.
SHARE:
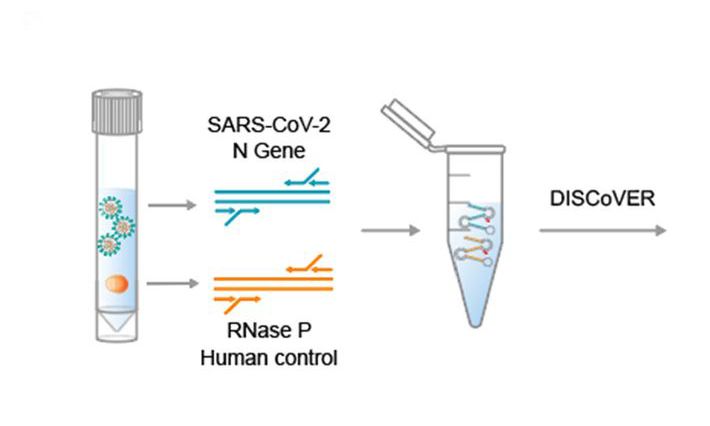
Conserved structural elements have already been shown to play critical functional roles in the life cycles of coronaviruses. Targeted disruption of regulatory functions of these structural elements, for example, through therapeutic targeting of associated RNA helicases, provides a largely unexplored strategy that can limit viral loads with minimal impact on the biology of normal cells. While this idea would have been far-fetched a mere five years ago, advances in computational modeling and high-throughput experimental RNA shape analyses have all but overcome the critical barriers.
Our goal is to apply these tools to the study of SARS-CoV-2, the virus that causes COVID-19, to systematically identify the RNA structural elements in its viral RNA and to reveal the RNA-binding proteins and RNA helicases that serves as the chaperones that shape and maintain the structure of these elements. Many RNA helicases are suitable drug candidates; therefore, targeting the RNA chaperons of SARS-CoV-2 is a viable therapeutic strategy to limit the spread or virulence of this virus.
This work is funded by the Laboratory for Genomics Research (LGR), a collaboration between UC Berkeley/UCSF (IGI) and GlaxoSmithKline.
Share this project: