Genomics
Institute
Manipulating chromatin to improve genome engineering
We are exploring how different chromatin domains in the eukaryotic nucleus can be manipulated to improve the efficiency of genome engineering.
SHARE:
Eukaryotic genomes are packaged into a spectrum of spatially and functionally distinct chromatin domains, represented at the extremes by transcriptionally-repressed heterochromatin and ‘active’ euchromatin. These different chromatin environments in the nucleus can negatively influence the efficiency of genome engineering and thereby limit its potential use.
Effective and efficient genome engineering relies on site-specific induction of DNA double strand breaks and repair by homology-dependent mechanisms. It is clear that both DNA damage induction and repair outcomes are influenced by chromatin composition, specifically by epigenetic modifications on histones and DNA. Our research using fruit flies has demonstrated that specific histone modifications and modifier proteins regulate the relative utilization of homology-dependent repair. However, we lack critical information about how specific chromatin modifications and modifier proteins influence the level of homology-dependent repair, as well as direct tests of the impact of manipulating chromatin states on genome engineering. Thus, our overarching goals are to 1) determine how chromatin composition impacts DNA damage repair 2) use this information to increase the frequency of homology-directed repair mechanisms critical for genome editing, and improve our ability to engineer ‘difficult’ chromatin regions, such as heterochromatin.
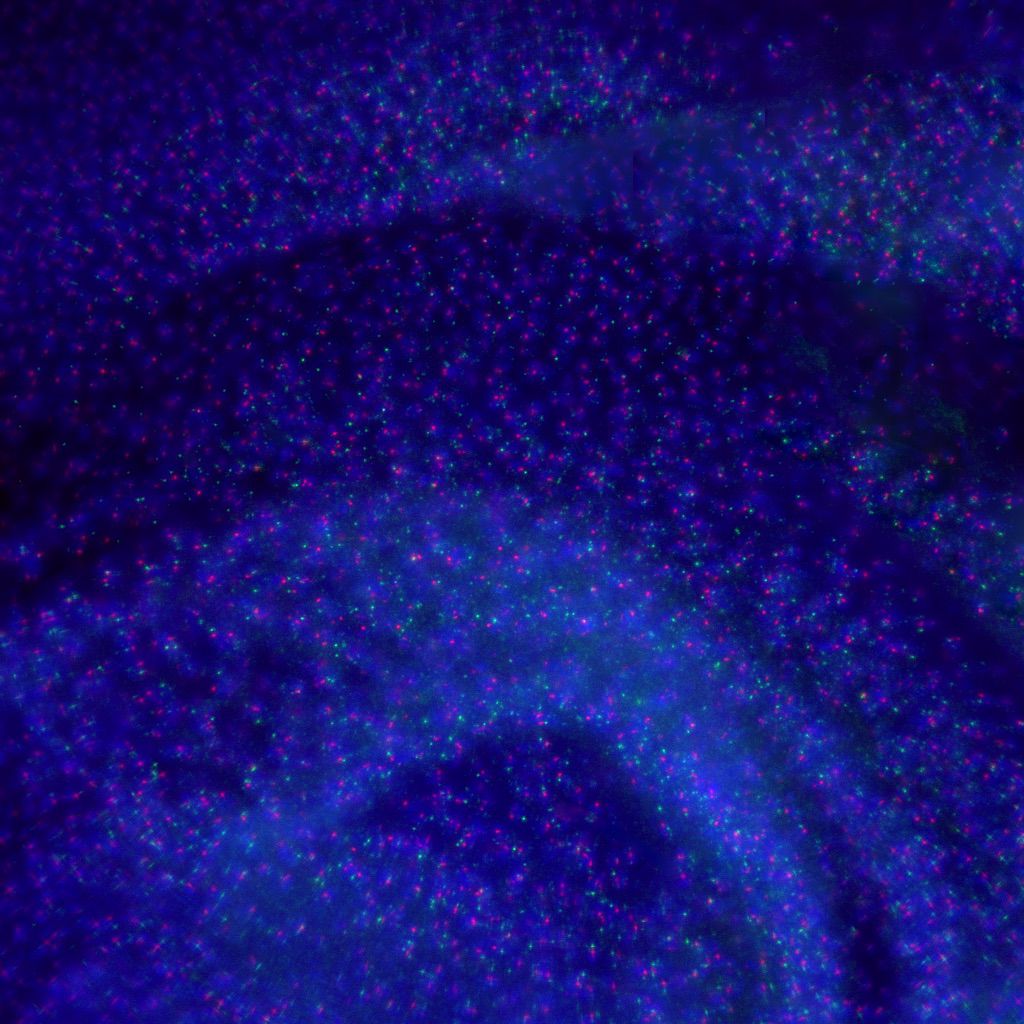
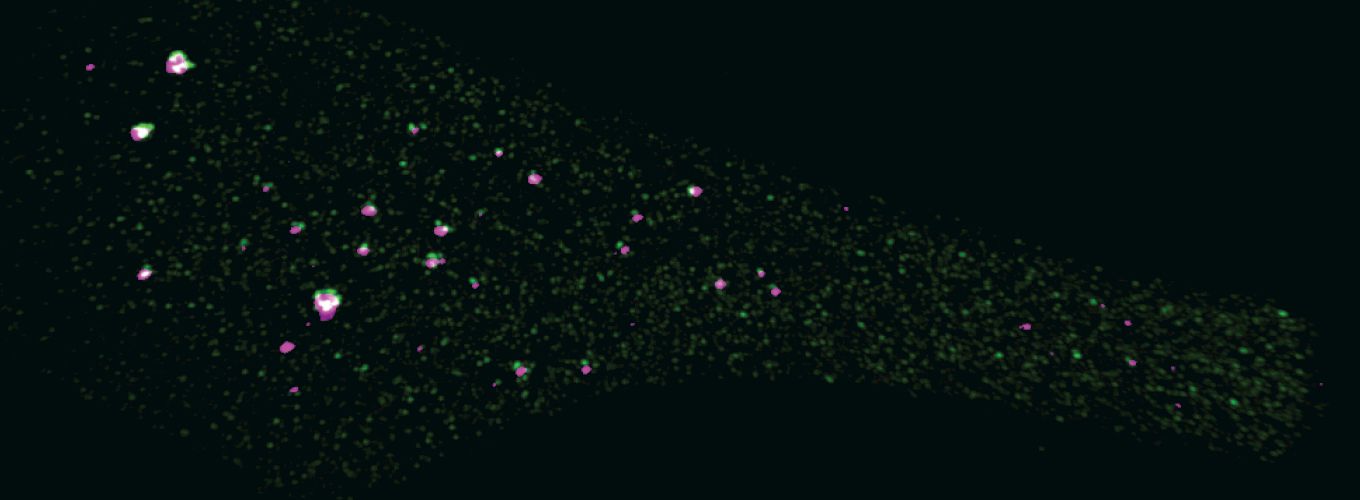
Photo credit: Aniek Janssen (Karpen Lab)

Photo credit: Aniek Janssen (Karpen Lab)
Photo credit: Aniek Janssen (Karpen Lab)
Photo credit: Aniek Janssen (Karpen Lab)
Share this project: