Genomics
Institute
Inhibiting COVID-19 (SARS-CoV-2) virulence protein function
We are developing a new approach to inhibit a key SARS-CoV-2 virulence protein using functional genomics and structural biology that can be adapted to future disease outbreaks.
SHARE:
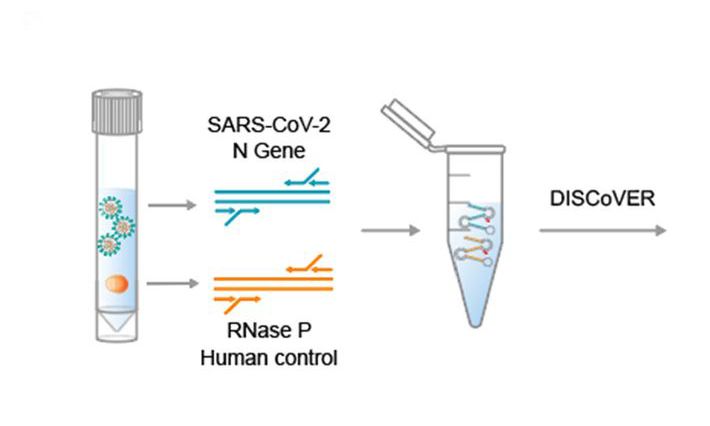
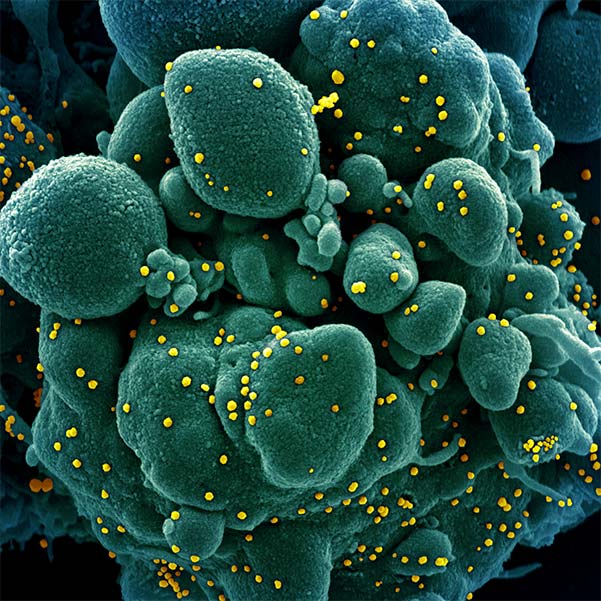
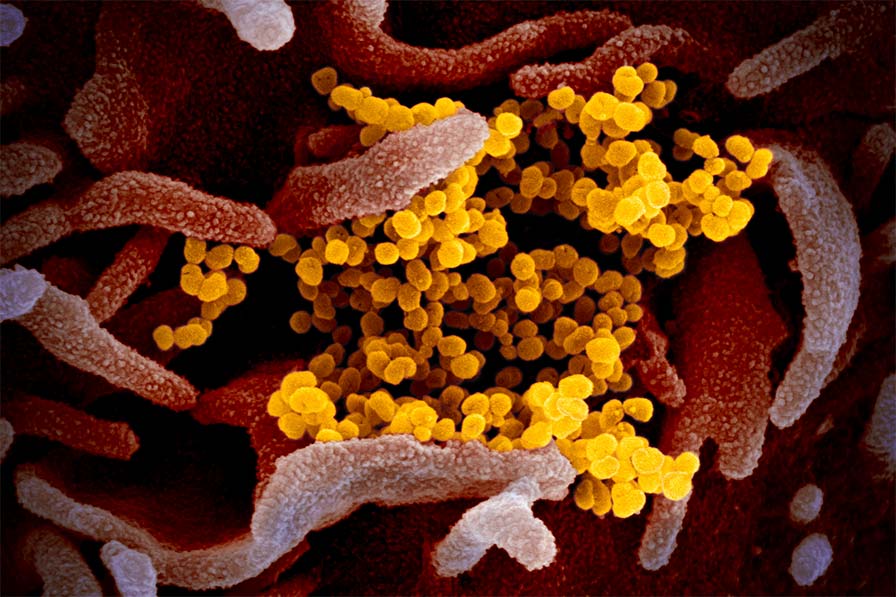
In the past 20 years, there have been three major beta-coronavirus (CoV) zoonotic transmission events and there are estimated to be over 5,000 additional coronavirus species circulating in bats. In each of these spillovers, the CoV-induced severe respiratory illness has been linked to delayed interferon signaling — allowing the virus to rapidly amplify — followed by subsequent acute lung injury, likely due to an inappropriately delayed and exuberant immune response. Underlying this ability to dampen host interferon signaling is the CoV nonstructural protein 1 (nsp1), a "host-shutoff" factor that effectively blunts expression of an array of host immune response genes.
Our goal is to provide comprehensive, foundational insight into nsp1 residues, and host factors and sequences essential for the function of this conserved SARS-CoV-2 virulence protein. This approach can form the basis for targeted drug discovery that can be leveraged to combat COVID-19 as well as future disease outbreaks.
This work is funded by the Laboratory for Genomics Research (LGR), a collaboration between UC Berkeley/UCSF (IGI) and GlaxoSmithKline.
Share this project: