Genomics
Institute
Improving photosynthesis in crop plants by editing chloroplasts
Project Overview
We are using CRISPR tools to increase the efficiency of photosynthesis.
Each year, five billion tons of carbon are added to the atmosphere. Meanwhile, over 100 billion tons of carbon are fixed every year through the Calvin Benson Bassham (CBB) cycle, the carbon-fixing pathway in photosynthesis. Rubisco, the most abundant enzyme on the planet, catalyzes the key step of the CBB cycle by adding one molecule of CO2 to a five-carbon sugar, forming two three-carbon sugars.
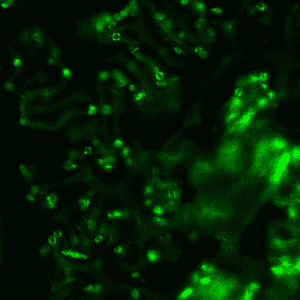
A chief barrier in efforts to improve photosynthesis in plants has been the technical difficulties associated with the genetic engineering of rubisco, as it is contained within the chloroplast genome. One challenge that must be overcome in order to achieve higher throughput in chloroplast transformation is selection of plant cells containing a single chloroplast genome (homoplasmy) and subsequent regeneration of a complete plant. Since plant cells typically contain hundreds of chloroplasts, each of which contains tens of copies of its genome, and chloroplast genomes typically segregate freely, achieving homoplasmy in the face of extreme polyploidy requires multiple rounds of strict selection.
We hope to create a novel selection system for the streamlined isolation of homoplasmic mutants. We plan to use a CRISPR-based approach to target unedited genomes for destruction, enriching genomes with the desired edit. If successful, this will enable more rapid, high-throughput testing of edits for the improvement of photosynthesis.