Since the development of CRISPR-Cas9 genome editing in 2012, dozens of Cas enzymes and variants have become available to researchers. The toolkit is always expanding, with enzymes that can offer more precision, smaller size, and even different functions. The Doudna lab at the IGI has created a comprehensive, free resource to help researchers pick which enzyme best fits their experimental needs: CasPEDIA.
CasPEDIA is a searchable, wiki-style database of Class 2 Cas enzymes, detailing properties including protein structure, nuclease activity, target requirements, previous uses, and intended applications. The database also introduces a novel functional nomenclature, CasIDs, inspired by the Enzyme Commission (E.C.) number system. CasIDs streamline the identification of nucleases by consolidating enzyme properties into a 3-decimal system.
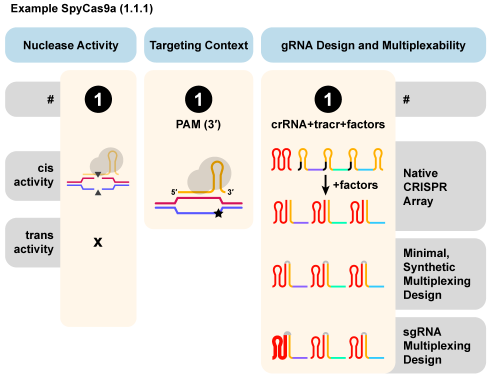
The Doudna lab team will continue to update the database and, like Wikipedia, users are encouraged to add entries themselves. Explore by clicking the button below!
Learn more about CasPEDIA:
- CasPEDIA: A Functional Classification of Cas Enzymes
- CasPEDIA Database: a functional classification system for class 2 CRISPR-Cas enzymes
Other IGI resources: