Advanced Translational Genetics Laboratory

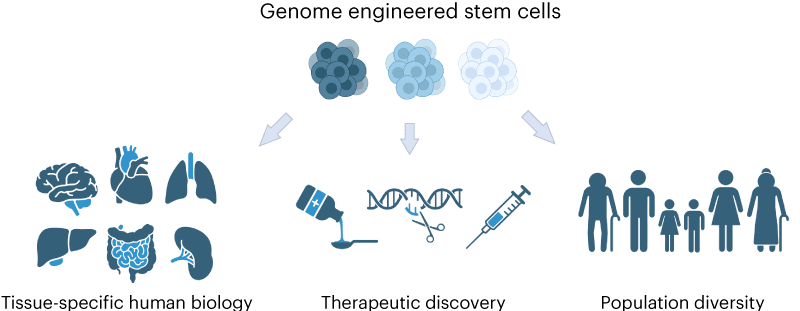
The laboratory for Advanced Translational Genetics (ATG) develops and deploys scaled stem cell engineering workflows to elucidate molecular principles of human disease and discover novel therapeutic strategies on diverse genetic backgrounds. Currently, the ATG focuses on research and development of novel models for cancer and neurodegenerative diseases.
The cornerstone of the ATG lab is an automated tissue culture system (ATTIS) that enables scaled genome editing projects to generate isogenic hPSCs carrying designer mutations. ATTIS is operated by a team of expert molecular biologists and automation engineers that streamlines the generation and analysis of the gene-edited hPSCs allowing a cell culture and editing pipeline that presents a throughput 20–50 times higher than currently accessible in manually conducted hPSC editing experiments.
The ATTIS in action in the ATG laboratory at the Innovative Genomics Institute
ATG Team

Hanqin Li, Ph.D.
P.I. Advanced Translational Genetics Laboratory

Tomy Tran
Automation Engineer


Aashaka Kalavadia
Research Associate

Henry Feng
Undergraduate Researcher

Jianpu Gao
Undergraduate Researcher

Ashley Heng
Undergraduate Researcher

Kim Lund
Undergraduate Researcher

Broc Vargo
Undergraduate Researcher
Collaboration
The ATG lab is now soliciting proposals from stem cell focused research teams for collaborative projects that would advance stem cell-based disease modeling at the Innovative Genomics Institute (IGI), UC Berkeley, and institutions and communities from the greater Bay Area.
All projects with ATG are scientific collaborations. We will work together with the applicant’s team on experimental design, workflow adaptation, material and data handling and budgeting. ATG will cover the operational costs to successfully complete the project. Applicants will be responsible for reagent and consumable costs. The goal for the ATG team is to collaborate on joint publications and protection of technological and scientific advances enabled by the ATG by joint patent filings as appropriate. Whenever possible we envision publishing in open source journals and making cell lines available to the research community as soon as possible after they are generated.
All proposals will be reviewed by a multidisciplinary review committee and evaluated based on the compatibility and availability of ATTIS and the ATG team, and the overall alignment with the IGI’s mission to improve human health. Previous experience with the handling of human pluripotent stem cells (induced pluripotent stem cells or human embryonic stem cells) is required. Projects are performed as collaboration and laboratories need to be able to adequately maintain the returned hPSCs, process the genetic material, and analyze the genetic data as it fits into the agreed upon collaboration.
Biannual call for the generation of individually edited hPSCs
When permitted by the system’s capacity, we will have an open call twice a year to support small-scale projects that require individually edited hPSCs. We hope to facilitate a wider adaptation of hPSCs models in human disease research and serve our local stem cell community.
During the project collecting period (March and September when the capacity allows), collaborators are welcome to submit the mutations (at most 2 per lab) they want to have as isogenic hPSCs. The ATG team will handle the entire genome editing process, including editing strategy design, tissue culture, and genotyping. We will cover the operational cost while the reagent cost will rely on the collaborator labs.
Publications
Functional annotation of variants of the BRCA2 gene via locally haploid human pluripotent stem cells
Li H, Bartke R, Zhao L, Verma Y, Horacek A, Ben-Natan AR, Pangilinan GR, Krishnappa N, Nielsen R, and Hockemeyer D. Nature Biomedical Engineering
Highly efficient generation of isogenic pluripotent stem cell models using prime editing
Li H, Busquets O, Verma Y, Syed KM, Kutnowski N, Pangilinan GR, Gilbert LA, Bateup HS, Rio DC, Hockemeyer D, and Soldner F. eLife
Contact Us
Students and researchers interested in working with molecular biology, bioautomation, and bioinformatics within the ATG lab, please contact hanqinli@berkeley.edu.
Connected Research Centers
The ATG Laboratory is a core unit of the IGI Center for Translational Genomics.
For more information, also see: