

New research shows how plants directly recognize pathogens, and how to help them fight off other attackers.
What do bananas, wheat, chocolate, and humans have in common? They are all in the midst of deadly pandemics. Humans have the tools to fight back; plants need help.
“There are three pandemics in the plant world right now around the world,” says Brian Staskawicz, Director of Sustainable Agriculture at the Innovative Genomics Institute, and Professor of Plant & Microbial Biology at UC Berkeley. “Bananas are being threatened by the Fusarium Tropical Race 4 fungus, which has now been found in Latin America and is a serious concern. A disease called ‘wheat blast’ has now popped up in Africa, and is threatening the world’s wheat and overall food security. A third is threatening a plant that is near and dear to everyone’s heart: chocolate.”

Roughly half of the world’s chocolate is grown in West Africa, and it’s being threatened by cacao swollen shoot virus, a pathogen transmitted by mealybugs that rapidly kills cacao plants. The most dire predictions foretell an unthinkable future without chocolate, one that might only be 10 years away.
In a new study published this week in the journal Science, IGI researchers made a critical step toward helping plants fight pathogens without pesticides or other toxins. Staskawicz with co-PI Eva Nogales and their team have discovered the structure and function of a resistosome, a plant immune receptor that recognizes pathogens and activates a strong defense. But just like in humans, plant immune responses aren’t perfect, and they don’t have effective weapons to combat to all pathogens. With this structure, the IGI team now has a roadmap to give plants new tools to fight back.
How plants defend themselves
Both plants and animals respond to pathogens with proteins called nucleotide-binding leucine-rich repeat receptors (NLRs) that detect the presence of pathogens and initiate some form of an immune response.
“All pathogens deliver proteins into their hosts. It’s these receptors that intercept those proteins and trigger resistance to the pathogen,” says Staskawicz.
The IGI team focused on a specific NLR receptor called ROQ1 from Nicotiana benthamiana, a type of tobacco plant in the nightshade family. In 1994, the Staskawicz lab was the first to isolate the DNA sequences for these types of NLR genes, but until now — 26 years later — no one had been able to figure out the structure of the molecule, which is key for understanding how the receptor recognizes the pathogen and triggers an immune response.
Solving the structural puzzle
Researchers have been trying to understand the structure of plant resitosomes since the 1990s, but they had to wait for the techniques to emerge that would make it possible.

“The method that we used is electron microscopy, a technique that has been around for a long time, but it’s only been in the last six or seven years that the methodology has improved enough to get atomic structures,” says Eva Nogales, co-PI on the study, co-director of the Cal-Cryo facility at QB3-Berkeley, and senior faculty scientist at the Lawrence Berkeley National Laboratory. “This is a large protein assembly, it has flexible regions, and it is not trivial to produce it in large amounts! So crystallization and more traditional methods just have not been able to handle it.”
“Not trivial” turns out to be an understatement. Typically, when researchers use electron microscopy, they use insect or bacterial cell cultures to artificially produce many copies of the molecule in question. In this case, first author Raoul Martin, a Ph.D. candidate in the Nogales lab, had to turn back to the tobacco plant itself because the cell culture methods repeatedly failed to make usable protein.
“The amount we would get out of tobacco leaves was so little that I couldn’t even measure the concentration using standard methods,” says Martin.
He had to develop a new protocol to try to maximize the chance of capturing a molecule suitable for imaging, and after 12 attempts, he managed to get just one that worked — and it worked beautifully.
Having sufficient amounts of the molecule for imaging is one hurdle; the next is being able to accurately capture an image of the structure. This was made possible by being at UC Berkeley: the Cal-Cryo facility has state-of-the-art cryo-electron microscopes for conducting high-resolution imaging experiments at extremely low temperatures. Facilities like this are only available in a few places around the world.
Nogales finds the ROQ1 structure especially interesting, not just for it’s biology, but for a reason even a non-biologist can easily understand: it’s beautiful.
“I have been working on imaging molecular structures for over 20 years, and I only remember one other time that I was so amazed by the beauty of a structure, and that was with Jennifer Doudna’s first CRISPR structure called ‘Cascade,’ which looks like a seahorse,” says Nogales.
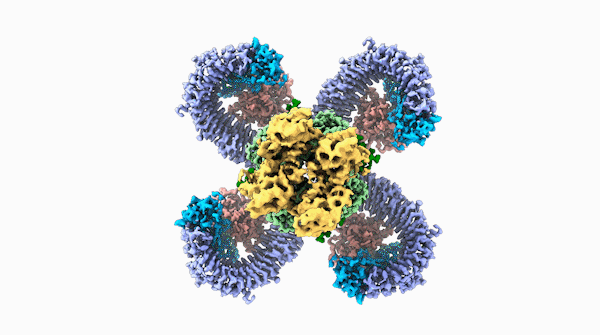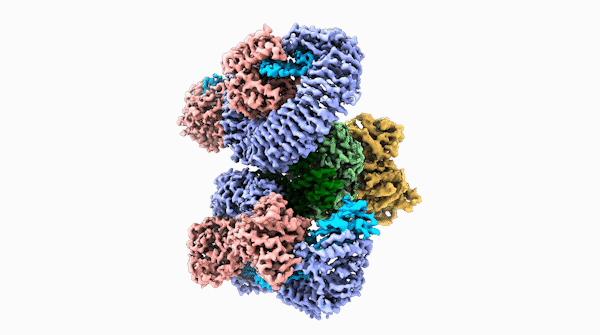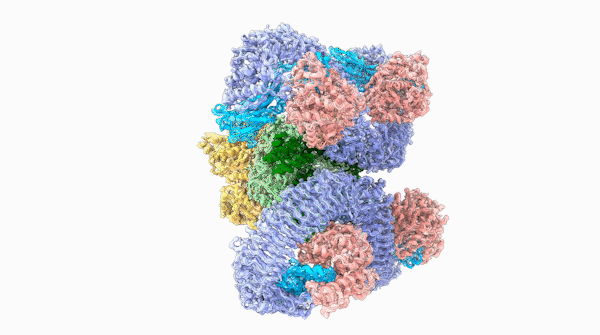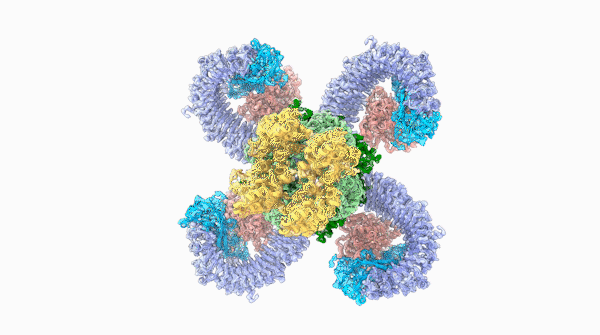
A striking similarity
For Raoul Martin, it wasn’t just the beauty of the molecule that stopped him in his tracks, it was that the structure looked oddly familiar. The way the loops of the ROQ1 molecule recognize pathogens was remarkably similar to antibodies that you find in mammals.

“I took a step back and said, ‘Oh, wait a minute,’” says Martin. “Every biochemist has seen the structure of an antibody. This is one of the classic things that you learn about, and I worked in a lab as an undergrad that studied them. As soon as I saw the structure, I thought, ‘Wow, that looks exactly like the folds I’ve seen in antibodies.’”
The structures are clearly an example of convergent evolution according to Martin. In other words, they didn’t derive from the same ancestor, but they arrived at a similar strategy to defend against attacks at the molecular level.
The team was able to show that loops on one end of the complex, cloverleaf-shaped molecule can directly recognize pathogen proteins, the first time this has been shown in plants. When this happens, the ROQ1 molecule transforms in a way that activates a defensive response in the plant tissue. In addition, the structure also seems to have a mechanism that directly inactivates the pathogen protein.
Engineering immunity
The similarity to mammalian antibodies isn’t just an interesting example of evolutionary symmetry, it suggests a course of action for the IGI team. Now that they understand how the plant resistosome recognizes pathogens using specific loops of the molecule, they know from similar work in animals that it’s possible to reengineer those loops to recognize — and hopefully fight off — other pathogens. That is, they can create new resistance genes designed to protect plants from specific attackers, like protecting bananas from Fusarium, wheat from wheat blast, and chocolate from cacao swollen shoot virus. Each plant system is different in the details, but the principles remain the same.
This finding raises an important evolutionary question: If the loops on resistosomes recognize specific pathogens, how did plants acquire resistance in the first place? And why are they lacking resistance to other pathogens?
“My feeling is that after we solve about a hundred of these structures, then we’ll be able to have a better theory,” says Staskawicz. “The more structures we have, the better off we’ll be.”
For Martin, this project was especially gratifying because his previous research project during his Ph.D. studies had failed. This project turned his doctoral research around completely.
“I can say this,” Martin jokes, “tobacco kills, but it saved my Ph.D.”
Read more:
Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ
Perspective: Enzyme formation by immune receptors
Media Contact
Andy Murdock: andymurdock@berkeley.edu



