Genomics
Institute
Molecular genetic analysis of the microbiome using CRISPRi knockdowns
We are building new tools to manipulate gut bacteria, allowing us to discover how bacteria interact with each other in communities, with pharmaceuticals, and with their human hosts.
SHARE:
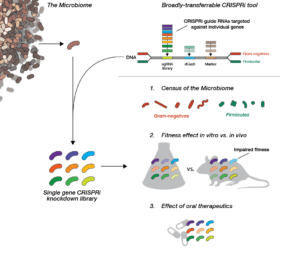
Mammals are home to complex microbial communities (microbiomes) that have a profound impact on host health and disease. However, our ability to systematically determine which microbial genes and processes are critical to microbiome function is severely limited by the absence of facile genetic tools for almost all of its constituent microbes. Indeed, of the nine most prevalent bacteria genera in the microbiome, only Bacteroides has been amenable to genome-scale transposon mutagenesis. There is a clear need for a scalable genetic technology that will be broadly applicable to the constituent strains of the microbiome. Programmable transcriptional repression mediated by a catalytically inactivated Cas9 (dCas9 from S. pyogenes) and a single guide RNA (sgRNA), a technology termed CRISPR interference (CRISPRi), has emerged as a flexible tool for genome-scale genetic analyses by allowing for repression of any gene with high precision. We are using our newly-developed, broadly-transferrable CRISPRi knockdown tools to establish CRISPRi as a foundational technology for genetic dissection of the gut microbiome by: (1) building and sharing molecular genomic resources for gut bacteria, (2) determining which genes contribute to the fitness of gut bacteria species in both in vitro culture and an in vivo mouse model, and (3) investigating how orally-administered therapeutics cause (unintended) antimicrobial activity.
Share this project: